Structural and functional studies of erythrocyte membrane-skeleton by single-cell and single-molecule techniques
Abstract
As the indispensable oxygen-transporting cells, erythrocytes exhibit extreme deformability and amazing stability as they are subject to huge reversible shear stress and extrusion force during massive circulation in the body. The unique architecture of spectrin-actin-based membrane-skeleton is considered to be responsible for such excellent mechanical properties of erythrocytes. Although erythrocytes have been recognized for more than 300 years, myriad questions about membrane-skeleton constantly attract people’s attention. Here, we summarize the kinds of distinctive single-cell and single-molecule techniques that were used to investigate the structure and function of erythrocyte membrane-skeleton at macro and micro levels.
1. Introduction
Erythrocytes are the largest number of blood cells that deliver oxygen from the lungs to the body tissues. The erythrocytes possess a unique biconcave-disk profile that provides excellent deformability and stability to survive the huge shear force and extrusion force during massive circulation in their life. It has been suggested that the characteristic architecture of erythrocyte membrane-skeleton is responsible for such extreme mechanical deformability.1,2 The organization of membrane-skeleton is rather complex, and current models depict that the membrane-skeleton is principally composed of a lipid bilayer with an attached skeleton formed by a two-dimensional triangular network of the spectrin tetramers linked by junctional complexes consisting of short actin filaments, protein 4.1, adducin, tropomodulin, tropomyosin and other associated proteins (Fig. 1).3–5 This membrane-skeleton plays an important role in maintaining the physiological functions of erythrocytes. It has been demonstrated that the deformability decrease resulted from membrane-skeleton defects underlie the generation of various diseases such as diabetes,6 anemia,7 spherocytosis,8 malaria infection,9 and so on.
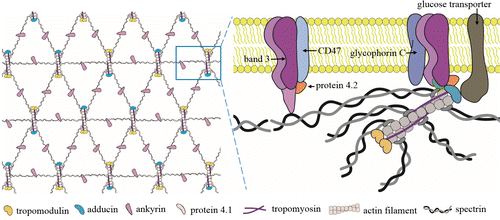
Fig. 1. Current model of the spectrin-actin-based structure of erythrocyte membrane-skeleton. A two-dimensional triangular meshwork composed of rod-like spectrin tetramers that connect at junctional complexes containing short actin filaments and associated proteins.
The considerable progresses in the knowledge of the structure and function of erythrocyte membrane-skeleton deeply rely on the application of advanced single-cell and single-molecule techniques. For instance, the optical tweezers,10 microfluidic chips11 and micropipette aspiration12 have emerged as potent methods for investigating membrane-skeleton-dependent deformability of erythrocyte at single-cell level. Meanwhile, the sub-cellular nanoscale organization of the membrane-skeleton has been well studied by electron microscopy (EM)13,14 and atomic force microscopy (AFM).15 Furthermore, multiple techniques including immuno-electron microscopy (iEM),16 antibody recognition imaging by AFM17 as well as super-resolution fluorescence microscopy (SRFM)18 provide new opportunities to uncover the ultrastructure of the erythrocyte membrane-skeleton at single-molecule level.
The present review focuses on the current understanding of the structure and function of erythrocyte membrane-skeleton based on single-cell and single-molecule level methods.
2. Membrane-Skeleton-Dependent Deformability of Erythrocyte Studied by Single-Cell Techniques
Devoid of nucleus and intracellular organelles, the extreme deformability of erythrocyte predominantly relies on its unique architecture of membrane-skeleton system. In other words, the erythrocyte deformability could directly and effectively reveal the membrane-skeleton properties. The better deformability corresponds to healthier organization of membrane-skeleton.
2.1. Optical tweezers
Optical tweezers, first achieved in 1986 by Ashkin who was rewarded Noble Prize in 2018, trapped and manipulated dielectric particles by laser beams based on radiation forces of pico-newton scale.19 As a powerful tool to manipulate atoms,20 particles21 and cells,22 optical tweezers are finding increasingly widespread applications in the investigation of various physical and biological processes. Specifically, optical tweezers have shown tremendous potential in measurement of erythrocyte deformability at single-cell level.
Early optical tweezers consisted of a single-beam optical trap formed by a laser to manipulate erythrocytes. Individual erythrocytes were stably suspended inside a flow chamber by application of a single-beam optical tweezers, where the bending rigidity of the erythrocytes was observed to infer the deformability.23 On the other hand, the erythrocyte deformability could be studied when single-beam optical tweezer dragged erythrocyte at a constant velocity (Fig. 2(a)).24 In addition, the ATP-dependent nanoscale membrane fluctuations, an indicator of local erythrocyte membrane-skeleton mechanical properties, have been investigated by various techniques including flickering spectroscopy, positive phase contrast microscopy and point dark field microscopy.25–27 Furthermore, application of single optical tweezer enabled a highly accurate measurement of the erythrocyte membrane fluctuation amplitudes with microsecond temporal and sub nanometer spatial resolution through laser scattering detection.28
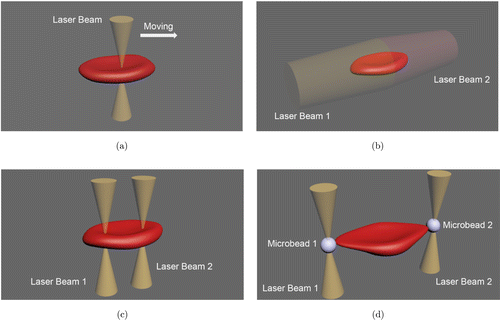
Fig. 2. Schematic of different optical tweezers on erythrocyte. (a) The erythrocyte deforms when moving with the single-beam optical tweezer. (b) The optical stretcher uses two nonfocused laser beams from opposite directions to deform the erythrocyte. (c) The dual-trap optical tweezer applies two parallel beams focused in the interior of the erythrocyte. (d) The laser beams stretch the erythrocyte through microbeads attached to the cell.
The optical stretcher, a variant of optical tweezers, uses two nonfocused laser beams from opposite directions to stably trap the erythrocyte (Fig. 2(b)). The erythrocyte deformability was assessed by shape change when it was stretched out along the laser beam axis.29 The radiation damage to the cells can be ignored for optical stretcher owing to the defocused beams, which minimizes the light flux through the cells in comparison to other optical traps using focused lasers.
Distinguishing from the optical stretcher with two counter-propagating-beams, the dual-trap optical tweezer uses two parallel trapping beams that focused in the interior of the erythrocyte. The erythrocyte is stretched when the two beams are separated by a distance (Fig. 2(c)). This developed optical tweezers provided precise control to the erythrocytes, and the stretched length of erythrocytes was measured to assess the deformability.30,31 Furthermore, a more complex optical tweezer with three optical traps was designed to manipulate erythrocytes from three dimensions simultaneously.32
In addition, optical tweezers can also manipulate erythrocytes through microbeads attached to them as shown in Fig. 2(d). Based on this technique, the erythrocyte deformability was indicated by the change of projected diameter of the erythrocyte.33 The elasticity of erythrocyte membrane-skeleton was investigated through applying force to beads attached to erythrocyte ghosts.34 The relaxation response of the erythrocytes stretched by this optical tweezer was also recorded to assess the deformability by the relaxation time after large deformation.35 This microbeads-based optical tweezer could induce larger deformation of erythrocytes than that of pure optical tweezers.
Diversified optical tweezers have been developed to study erythrocyte deformability in different conditions including malaria infection,36 sickle cell disease,37 osmolality change38 and trapping in living animals.39 It found that erythrocyte deformability underwent a decrease in pathological conditions.36,37 These works may contribute to an in-depth understanding of the mechanical properties of erythrocyte membrane-skeleton.
2.2. Micropipette aspiration
Since the first use of micropipette aspiration for measuring the elastic properties of sea urchin eggs in 1954 by Mitchison and Swann,40 this method has been widely used in many types of cells, such as neutrophil cells,41 endothelial cells42 and chondrocyte cells.43 In particular, the micropipette aspiration permits quantitative analysis of the erythrocyte deformability.44
Depending on the diameter of the micropipette and the aspiration pressure, there are three types of erythrocyte aspiration, including aspiration of a small hemispherical portion of membrane-skeleton (Fig. 3(a)), an aspired tongue within the micropipette (Fig. 3(b)), and an aspiration of the whole cell into the micropipette (Fig. 3(c)). When a small hemispherical portion of the erythrocyte was aspired into the micropipette, the erythrocyte deformability was assessed by the surface elastic moduli, which could be calculated through the size of the aspired portion and the applied pressure.45,46 When erythrocyte was aspirated into a micropipette with a long tongue, the shear modulus of membrane-skeleton could be determined by the tongue length.47 The actin filaments of the membrane-skeleton were found to be obviously modified when erythrocyte was aspirated with a long tongue inside the micropipette.48 When the suction pressure increased large enough, the aspirated erythrocyte folded into the micropipette, in which case the bending elastic modulus could be derived from the applied pressure.49

Fig. 3. Schematic of micropipette aspiration on erythrocyte. (a) Aspiration of a small hemispherical portion of erythrocyte membrane-skeleton. (b) An aspired tongue of erythrocyte membrane-skeleton within the micropipette. (c) An aspiration of the whole erythrocyte into the micropipette.
In comparison with optical tweezers, the micropipette aspiration could supply a local deformation on the erythrocyte membrane-skeleton. Meanwhile, the mechanical properties of the membrane-skeleton can be quantitatively revealed when the micropipette aspiration method combines with theoretical models.
2.3. Microfluidic channels
The microfluidic channels have also been demonstrated as a potent and useful method for investigation of erythrocyte deformability. Various parameters on the morphology of erythrocytes in microfluidic channels can be quantified to assess the deformability. For instance, deformation index (DI=L∕DDI=L∕D, LL is the height, and DD is the width of the parachute, Fig. 4(a)) was defined to measure erythrocyte deformation when they folded into a parachute-like shape in the microfluidic channels.50,51 The cortical tension, another indicator of the deformability, could be calculated by the deformed erythrocytes when they extrude through a microchannel with repeated funnel pores.52,53 The transit velocity of the erythrocyte in the microfluidic channels was often used to assess the deformability. In detail, a microchannel with repeated taper constrictions was designed to detect deformability difference between healthy and P. falciparum-infected erythrocytes based on transit velocity measurement (Fig. 4(b)).54 A microchannel with periodically triangle-shaped pillars was fabricated to study the deformability of iron dextran-treated erythrocyte through the transit velocity calculation.55 A narrow straight microfluidic channel was applied to investigate the effects of drugs-induced actin filaments assembly/disassembly on erythrocyte deformability determined by alterations in cell transit time.56
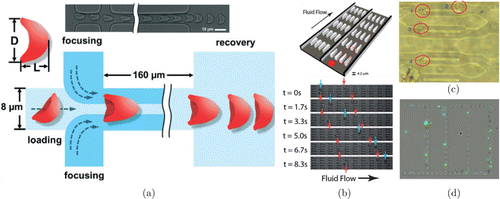
Fig. 4. The erythrocyte deformability studied by microfluidic channels. (a) Schematic of a microfluidic device for studying erythrocyte deformability by deformation index. (b) The microchannel with repeated taper constriction characterizes the erythrocyte deformability based on transit velocity detection. (c) The multi-channel microfluidic network simulates the blockage of the microcirculation inside capillaries. (d) The biomimetic microfluidic chip simulates the human spleen slits that capture poorly deformable erythrocytes. (a) is adapted with permission from Ref. 51, (b) is adapted with permission from Ref. 54, (c) is adapted with permission from Ref. 60 and (d) is adapted with permission from Ref. 61.
Furthermore, microfluidic channels possess a unique ability to simulate capillary microcirculation system in vitro,57,58 especially the blockage of capillaries. For example, the behavior and morphology of poorly deformable erythrocytes in capillary blockages were simulated and described using microchannels with different width.59 The degradation of the erythrocyte deformability was measured through the applied pressure and flow rate inside the multi-channel microfluidic network (Fig. 4(c)).60 In addition, the deformability of erythrocytes was characterized through the capture status in a biomimetic microfluidic chip with mechanical constraints that was designed to simulate the human spleen slits (Fig. 4(d)).61
Based on application of different microfluidic channels, they all suggested that erythrocyte deformability decreased significantly under various abnormal conditions, such as malaria-infection,62,63 blood storage lesion,64,65 diabetes66,67 and sickle cell disease.68,69 Compared with the optical tweezers and the micropipette aspiration, the microfluidic channels provided a unique opportunity to investigate erythrocyte deformability in a rapid, high-throughput and relatively easy-to-use method at single-cell level.
3. Ultrastructure of the Erythrocyte Membrane-Skeleton Studied by Single-Molecule Techniques
3.1. Immuno-electron microscopy (iEM)
Taking advantage of the ultrahigh imaging resolution of EM, the sub-cellular ultrastructure of erythrocyte membrane-skeleton has been primarily described by different kinds of EM.13,14,16,70–72 Early results from negative staining EM (Fig. 5(a)),70 as well as quick-freezing, deep-etching and rotary-replication (QFDERR) (Fig. 5(b))16 clearly showed that the erythrocyte membrane-skeleton formed a dense triangular network of intersecting straight filaments linked by rod-like junctional complexes at the converging nodes at sub-cellular level. Interestingly, the junction-to-junction distances of the triangular network were ∼∼200nm in spread erythrocyte membrane-skeleton13 and 30–40nm in nonspread membrane-skeleton.72 Furthermore, a work used cryo-electron tomography to evaluate the three-dimensional topology in intact, unexpanded erythrocyte membrane-skeleton, which revealed a complex filament meshwork with an average edge length of 46nm (Fig. 5(c)) and a gradual decrease in both the density and the thickness of the network from the center to the edge of the cell at sub-cellular level.14 Recently, cryo-electron tomography was used to reveal the remodeling of actin filaments in Plasmodium falciparum-infected erythrocytes.73,74
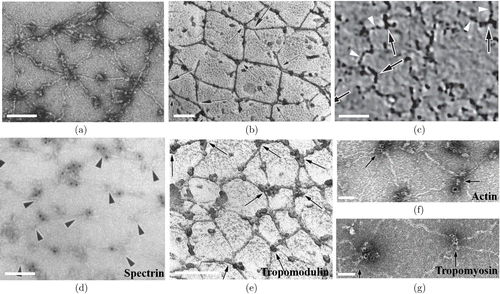
Fig. 5. The ultrastructure of the erythrocyte membrane-skeleton shown by different kinds of EM. (a) Spread erythrocyte membrane-skeleton imaged by negative-staining EM. (b) An unfixed erythrocyte membrane-skeleton visualized by QFDERR. (c) Cryo-electron tomography of the intact, unexpanded membrane-skeleton showing a complex filament meshwork. (d) Immunogold labeling of spectrin in human erythrocyte membrane-skeleton visualized by negative staining. (e) Immunogold labeling of TMOD in erythrocyte membrane-skeleton obtained by QFDERR. (f, g) Immunogold labeling of actin (f) and tropomyosin (g) in erythrocyte membrane-skeleton visualized by negative staining. Scale bar==100 nm. (a) is adapted from Ref. 70, (b) and (d) are adapted from Ref. 16, (c) is adapted from Ref. 14 and (e) (f) and (g) are adapted from Ref. 78.
The development of iEM, which combines immunogold labeling with above EM techniques, enables the single-molecule measurements of erythrocyte membrane-skeleton architecture.16,72,75 Conventional electrophoresis and biochemical analysis only showed that erythrocyte membrane-skeleton contains ∼∼20 major proteins such as spectrin, actin, tropomodulin, band 3, protein 4.1, ankyrin, etc. without actual spatial distribution information.76,77 Labeling the membrane-skeleton with gold particles that coated with site-specific spectrin antibodies verified that the filaments in the meshwork were spectrin molecules based on negative staining EM (Fig. 5(d))16 and QFDERR.72,75 Immunogold labeling of band 3 molecules further demonstrated that integral membrane proteins were organized into macromolecular complexes centered on band 3 protein, which were anchored to the membrane-skeleton.72 Moreover, immunolocalization of tropomodulin (Fig. 5(e)), actin (Fig. 5(f)) and tropomyosin (Fig. 5(g)) in spread erythrocyte membrane-skeleton provided direct evidence that short filaments at the central junctions were indeed actin filaments, and both tropomodulin and tropomyosin were associated with the actin filaments.78 iEM could distinguish normal biconcave erythrocytes from abnormally-shaped erythrocytes obtained from patients by distinct patterns of spectrin filaments.79
3.2. Atomic force microscopy (AFM)
Compared with EM, normal AFM has the advantage of in situ imaging and high sensitivity to small height variations in surfaces at sub-cellular level, thereby it has been widely used in measuring the sizes, shapes and mechanical properties (such as stiffness, viscoelasticity, hardness and adhesion) of erythrocyte.80–82 AFM nano-mapping revealed that pathological erythrocytes usually lost the unique biconcave-disk shape and exhibited decreased membrane roughness as well as increased membrane stiffness compared with healthy erythrocytes.82–84 Furthermore, AFM could examine the nanoscale ultrastructure of the erythrocyte membrane-skeleton. For instance, the topography of the membrane-skeleton on the cytoplasmic surface of the erythrocyte plasma membrane was directly observed in freeze-dried ghosts at the external surface of the cell without removing the membrane or extending the membrane-skeleton.17 Additionally, another work demonstrated the applicability of AFM for imaging the two-dimensional membrane-skeleton of untreated as well as fixed human erythrocytes under near physiological conditions (Fig. 6(a)).15
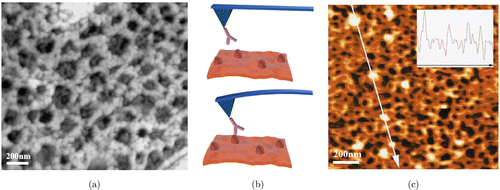
Fig. 6. The ultrastructure of the erythrocyte membrane-skeleton studied by AFM. (a) Two-dimensional overhead view of contact AFM image of a ghost taken from osmotically lysed and air dried unfixed erythrocytes. (b) Schematic of the principle of antibody recognition imaging by AFM. (c) An AFM image of a ghost membrane-skeleton immunogold labeling with spectrin antibodies. The line indicates the place where the cross section was measured, and passes four particles that are likely to be gold particles. The insert curve is the surface contour along the line. (a) is adapted with permission from Ref. 15 and (c) is adapted with permission from Ref. 17.
The correlation of AFM with single-molecule recognition imaging (also known as topography and recognition imaging, TREC), in which the AFM tip is tethered with specific molecules to recognize the counterpart molecules on the surface (Fig. 6(b)), expands the single-molecule level applications of AFM.85,86 These recognition pairs can be antigens and antibodies, hemagglutinin and oligosaccharides, ligands and receptors, enzymes and substrates, etc. Applying this combined method, Takeuchi et al. immunogold labeled spectrin with specific antibodies and demonstrated that the three-dimensionally folded meshwork was indeed the spectrin network (Fig. 6(c)).17
In addition, single molecule force spectroscopy (SMFS), a powerful approach derived from TREC imaging with AFM, can study protein interactions at the single-molecule level by detecting tens of pico-newton forces. Based on SMFS, Wang et al. observed the membrane proteins of erythrocyte and proposed a novel semi-mosaic model for the erythrocyte membrane organization.87–89 Although application of SMFS sheds new light on the studies of erythrocyte membrane proteins, the anatomy of erythrocyte membrane-skeleton at single-molecule level by AFM still lack a satisfactory level of understanding up to date.
3.3. Super-resolution fluorescence microscopy (SRFM)
According to the results of EM and AFM, it is remarkable to note that major discrepancies exist for the length of spectrin tetramers of the membrane-skeleton.13–17 Above studies had difficulty in obtaining the native ultrastructure of the erythrocyte membrane-skeleton due to the extensive sample processing, in which samples were often dried and/or membrane removed. Recently, the rise of SRFM provides new opportunities to visualize actual cellular ultrastructure with nanoscale resolution at single-molecule level with minimal sample damage.90,91
Three-dimensional stochastic optical reconstruction microscopy (3D-STORM) has been demonstrated as a powerful SRFM to investigate cytoskeletal biology.92–94 Our recent work revealed the native ultrastructure of the erythrocyte membrane-skeleton in membrane-preserved cells based on 3D-STORM (Fig. 7).18 We achieved molecular specificity for six targets, namely the N- and C-termini of ββ-spectrin, protein 4.1, F-actin, tropomodulin (TMOD) and adducin. We first developed an alternative method, in which live erythrocytes were first allowed to adhere to a polylysine-coated coverslip before subsequent fixation and labeling (Fig. 7(a)). It showed that the bottom membrane-skeleton was flat down to the axial resolution limit, which allowed us to obtain optimal 3D-STORM image quality (Figs. 7(b) and 7(c)). Based on STORM data of N-termini of ββ-spectrin, we revealed a ∼∼80nm junction-to-junction distance depending on the analysis of the distance between nearest neighbors (Figs. 7(d) and 7(e)) and two-dimensional autocorrelations (Figs. 7(d) and 7(e)), a length in agreement with relaxed spectrin tetramers under equilibrium,95 as well as theoretical predictions based on the abundance of spectrin in erythrocytes,96 but contrasting sharply with previous results from dried or membrane-removed erythrocytes by staining EM (∼∼200nm) and cryo-electron tomography (∼∼46nm),13,14 as well as from neurons under similar super-resolution settings (∼∼190nm).93 Interestingly, though presented at lower densities, statistics of distances between nearest neighbors still yielded peaks at ∼∼80nm for actin filaments and actin-capping proteins, TMOD and adducin, indicating that the actin-related targets occupy a subset of the same junctional complexes. Furthermore, cross-correlation calculation between N- and C-termini of ββ-spectrin indicated a ∼∼80nm junction-to-junction distance (Figs. 7(f)–7(h)). Meanwhile, we found that the dense erythrocyte membrane-skeleton often contained nanoscale voids (Figs. 7(f) and 7(g)), which may behave as structural weak points to facilitate fast changes of the erythrocyte shape during huge circulation. In addition, cross-correlation calculation between the protein 4.1 and TMOD showed a maximum value at zero intermolecular distance (Figs. 7(i)–7(k)), thus confirming co-localization at the nanoscale suggested from the in vitro interactions of purified proteins. In brief, our STORM data thus call for both experimental and theoretical reassessments of the structure and function of the native erythrocyte membrane-skeleton. Although our results could reveal the native organization of the erythrocyte membrane-skeleton depending on much more careful data analysis, it must admit that the STORM images fail to show direct visual pattern of membrane-skeleton as seen by the EM and AFM.
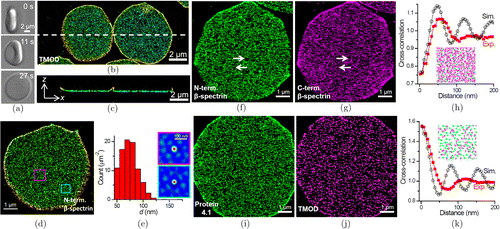
Fig. 7. The native ultrastructure of the erythrocyte membrane-skeleton revealed by super resolution fluorescence microscopy. (a) Differential interference contrast microscopy montages recording the bottom-flattening process of a live erythrocyte. (b) 3D-STORM picture of TMOD in bottom-flattened erythrocytes. (c) Virtual cross-section of the 3D-STORM image in the xzxz plane along the white dash line in (b). (d) 3D-STORM picture of the N terminus of ββ-spectrin. (e) Analysis of distances between nearest neighbors of ββ-spectrin clusters in (d). Insets: two-dimensional autocorrelation for the cyan- and magenta-boxed regions in (d). (f), (g), (i) and (j) STORM images of the N terminus (f), C terminus (g) of ββ-spectrin, protein 4.1 (i) and TMOD (j), respectively. (h) and (k) Two-dimensional pairwise cross-correlation calculation between the two color channels at different intermolecular distances based on the experimental (red) and simulated (black) data. Figures are adapted with permission from Ref. 18.
Recently, Huff from Zeiss Company developed a new form of super-resolution microscopy named AiryScan confocal fluorescence microscopy.97 Smith et al. used this super-resolution microscopy to image nonmuscle myosin II (NMII) motors in intact erythrocytes.98 Results indicated that NMII could form bipolar filaments, which associate with membrane-skeleton F-actin. Furthermore, NMII motor activity modulates interactions with the spectrin-F-actin network to control erythrocyte biconcave disk profile and deformability. This work provided a previously undescribed mechanism for actomyosin regulation of membrane tension, curvature, cell shape and biomechanics.
4. Conclusion
In conclusion, this paper summarizes the current understanding of the structure and function of erythrocyte membrane-skeleton based on kinds of distinctive single-cell and single-molecule level techniques. Each technique we mentioned has its advantages and also disadvantages, thereby making the integrated application of various techniques, such as correlation of EM and SRFM, to be helpful for researchers in further investigation of the unanswered questions underlying the structure and function of erythrocyte membrane-skeleton.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
Fulin Xing and Fen Hu contributed equally to this paper. This work was supported by the National Natural Science Foundation of China (Nos. 11874231, 11574165 and 31801134), Tianjin Natural Science Foundation (No. 18JCQNJC02000), the PCSIRT (No. IRT_13R29), and the 111 Project (No. B07013).