Algorithm to identify circulating tumor cell clusters using in vivo flow cytometer
Abstract
Recent studies in oncology have addressed the importance of detecting circulating tumor cell clusters because circulating tumor cell clusters might survive and metastasize more easily than single circulating tumor cells. Signals with larger peak widths detected by in vivo flow cytometer (IVFC) have been used to identify cell clusters in previous studies. However, the accuracy of this criterion might be greatly degraded by variance in blood flow and the rolling behaviors of circulating tumor cells. Here, we propose a criterion and algorithm to distinguish cell clusters from single cells. In this work, we first used area-based and volume-based models for single fluorescent cells. Simulating each model, we analyzed the corresponding morphology of IVFC signals from cell clusters. According to the Rayleigh criterion, the valley between two adjacent peak signals from two distinguishable cells should be lower than 73.5% of the peak values. A novel signal processing algorithm for IVFC was developed based on this criterion. The results showed that cell clusters can be reliably identified using our proposed algorithm. Intravital imaging was also performed to further support our algorithm. With enhanced accuracy, IVFC is a powerful tool to study circulating cell clusters.
1. Introduction
Circulating tumor cells (CTCs) have been considered an important biomarker for tumor diagnosis and prognosis.1,2 Recent studies in oncology have emphasized the importance of detecting circulating tumor cell clusters.3,4,5 These results indicated that circulating tumor cell clusters might survive and metastasize more easily than individual circulating tumor cells. The ability to detect circulating tumor cell clusters is necessary for studies of tumor metastasis. However, conventional in vitro detection methods are insufficient.6,7,8,9 The circulating tumor cell clusters may not be present in blood samples due to limited blood sample volumes. In addition, blood sample treatments may alter the properties of circulating cell clusters.10,11 For example, the addition of an anticoagulant may disperse the cell clusters, which may be a reason that little attention has previously been paid to circulating cell clusters.
The in vivo flow cytometer (IVFC) has been used to detect and enumerate CTCs in many reported studies.12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32 With longer detection times in vivo, a larger volume of blood is examined, and CTCs are more likely to be detected with IVFC compared to conventional in vitro detection methods. To accurately identify these cell clusters, we propose a new signal processing criterion and algorithm. Signals with larger peak widths, as detected by IVFC, have been used as a criterion to identify cell clusters in previous studies.3 However, the accuracy of this criterion might be greatly degraded by changes in blood flow and the rolling behaviors of circulating tumor cells.
Here, we propose a criterion and algorithm to distinguish cell clusters from single cells, based on computer simulation and intravital imaging. In this work, we established two models of fluorescent molecule distribution for two kinds of fluorescently labeled cells. To better analyze the signals from cell clusters, we first used area-based and volume-based models for single fluorescent cells. Using simulations of each model, we analyzed the morphology of IVFC signals from single cells and cell clusters. According to the Rayleigh criterion, the valley between two adjacent peak signals, from two distinguishable cells, should be lower than 73.5% of the peak values, and a novel signal processing algorithm for IVFC was developed based on this criterion. Our results showed that cell clusters could be reliably identified using our proposed algorithm. The results from intravital imaging also supported the use of our method.
2. Materials and Methods
2.1. Cell preparation
The 4T1 breast cancer cells were transfected with green fluorescent protein (GFP). They were cultured and expanded in an incubator, which maintained a temperature of 37∘C and 5% CO2. The cells were used after the sixth passage with stable GFP expression and then the cells were prepared for the simulation using the volume-based cell model.
2.2. Animal preparation
Balb/c mice (20±220±2g, 5 week-old) were purchased from Shanghai SLAC Laboratory Animal Co. Ltd. The animal treatment procedures were approved and monitored by the Ethical Committee of Animal Experiments in the School of Biomedical Engineering, Shanghai Jiao Tong University. The mice were anesthetized with 1% pentobarbital sodium salt (0.01mL/g mouse weight) during the experiment. To remove hair on the mouse ear, depilatory cream (sensitive hair removal cream; Veet) was topically applied.
2.3. IVFC
To simulate the proposed cell model, IVFC was used to collect signals from GFP-labeled cells. The schematic of IVFC was shown in supplementary Fig. S1. Briefly, the mouse was anesthetized and placed on the sample stage. With a 535nm LED as the light source, live images of the mouse’s ear were captured. These images were used to guide navigation of the laser beam onto a blood vessel for detection. Typically, 50–70μμm-wide blood vessels were selected as the detection site. The 488nm laser beams were modulated into slit-shaped beams using cylindrical lens. The size of the laser slit was approximately 5×725×72μμm at the focal plane. Under the guidance of the ear imaging by CCD camera, the laser slit was placed across the selected blood vessel. Fluorescently labeled cells were introduced into the blood by tail vein injection. When a cell passed through the laser beam, fluorescence was monitored by the detection system. After photo-electrical conversion via a photomultiplier tube (PMT), the signals were digitized (100KHz) and recorded on a computer.
2.4. Computer simulation and data processing
Computer simulations were performed on the MATLAB (2015a, Mathworks) computing platform. The plots were generated according to the area-based cell model or volume-based cell model. Data processing was performed according to the criterion and algorithm proposed using homemade MATLAB scripts.
2.5. Intravital imaging
Intravital imaging was performed with a Leica DM5500 system with a 10×× objective (water immersion, NA=0.6NA=0.6). The frame rate was 30fps.
3. Results
3.1. Area-based model of single cell
To better analyze a situation where multi-cells pass through the laser slit, models for single cells were first constructed. Two different models for single cells corresponded to different fluorescent labeling methods and fluorescent dyes.
The area-based single cell model was studied first. This model could be applied to fluorescent dyes that attached surface membrane of the cells, such as DiD. To simplify the simulation model, the laser power within the laser slit beam was considered uniform. The emitted fluorescence was proportional to the area that was irradiated by the laser beam.
In this model of peak signals, key parameters included the cell radius (R)(R), width of the excitation laser (W)(W), flow speed of the cells (vv) and time (tt). When the cell size was larger than the width of laser beam, the relationship between the parameters was as follows: R≥12WR≥12W. The process of the cell passing through the laser beam was divided into three phases (Fig. 1(a)). The relationship between fluorescence (F)(F) and tt during the whole procedure is plotted in Fig. 1(b).
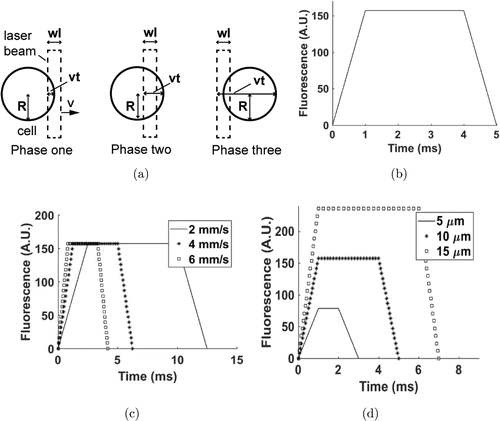
Fig. 1. Area-based single cell model. (a) Phases of cell passing the laser beam in area-based model. (b) Typical area-based model of single cell. In this model, R=10R=10μμm, W=5W=5μμm, v=5v=5mm/s and k=1k=1mV/μμm2. (c) Area-based model of single cell with different velocities. In this model, R=10R=10μμm, W=5W=5μμm and k=1k=1mV/μμm2. (d) Area-based model of single cell with different radiuses. In this model, W=5W=5μμm, v=5v=5mm/s and k=1k=1mV/μμm2. A.U. denotes arbitrary unit.
For the first phase, the front of the cell reached the front of the excitation laser beam, but the front of the cell had not reached the back of the excitation laser beam. This section was described as follows: 0≤vt≤W0≤vt≤W. The fluorescence (F)(F) detected within this time was described as follows :
For the third phase, the back of the cell passed the front of the excitation laser beam, but the back of the cell had not reached the back of the excitation laser beam. This section was described as 2R≤vt≤2R+W2R≤vt≤2R+W. The excitation area of the cell equaled πR(2R+W−vt)πR(2R+W−vt). The fluorescence (F)(F) over time was described as follows :
3.2. Volume-based model of single cell
The volume-based peak model indicated that the intensity of the emitted fluorescence was proportional to volume of cells excited by the laser. In this model, fluorescence peaks corresponded to cells expressing fluorescence proteins in the cytosol, such as GFP.
In this model, key parameters included the cell radius (R)(R), width of the excitation laser (ll), flow speed of the cells (vv) and time (tt). When cells were larger than the excitation laser area, the relationship between the parameters was as follows: R≥12WR≥12W. Based on the relative positions of the cell and excitation laser beam, the cell passed through the excitation laser in three phases (Fig. 2(a)). The relationship between FF and tt during this process is plotted in Fig. 2(b).
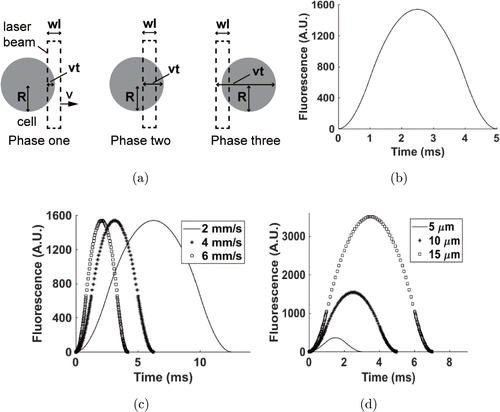
Fig. 2. Volume-based single cell model. (a) Phases of cell passing the laser beam in volume-based model. (b) Typical volume-based model of single cell. In this model, R=10R=10μμm, W=5W=5μμm, v=5v=5mm/s and k=1k=1mV/μμm2. (c) Volume-based model of single cell with different velocities. In this model, R=10R=10μμm, W=5W=5μμm and k=1k=1mV/μμm2. (d) Volume-based model of a single cell with different radii of cells. In this peak model, W=5W=5μμm, v=5v=5mm/s and k=1k=1mV/μμm2. A.U. denotes arbitrary unit.
For the first phase, the front of the cell reached the front of the excitation laser beam, but the front of the cell had not reached the back end of the excitation laser beam: 0≤vt≤W0≤vt≤W. Here, the excitation volume of the cell is the volume of the ball crown that is intercepted by the front end of the excitation laser beam. The fluorescence (F)(F) during this time is
For the second phase, the front of the cell had passed the back of the excitation laser beam, but the back of the cell has not reached the front of the excitation laser beam. This section is described as follows: W≤vt≤2RW≤vt≤2R. Here, the excitation volume in the cell equals the difference between the volumes of two ball crowns intercepted by the two ends of the excitation laser beam. After integrating, the fluorescence (F)(F) within the time was as follows :
For the third phase, the back of the cell has passed the front of the excitation laser beam, but the back of the cell has not reached the back of the excitation laser. This is described as follows: vv, tt and ll:2R≤vt≤2R+W2R≤vt≤2R+W. In the third phase, the excitation volume in the cell equals the volume of the ball crown intercepted by the back of the excitation laser beam. After integrating, the fluorescence (F)(F) within this time was as follows :
3.3. Models of dual-cell cluster
To better analyze the complex cases when clusters with multiple cells pass through the excitation laser beam, models of dual-cell clusters were designed, based on the single cell models above. For the dual-cell models, additional parameters were added, including the radii of the two cells (R1(R1 and R2)R2) and the distance between the front ends of the two cells at the horizontal axis (d). The horizontal axis corresponded to blood flow in the vessels. The distances are all referred to base on this horizontal axis. If the distance between the two cells was larger than the sum of both radii and the slit, the resulting peaks would not overlap in the time domain. This condition was the same as that of two separate single peaks. The relative sizes and positions of the two cells is the key concept here.
For the first cell, the process was similar to that in the single cell model. For the second cell, the starting time changed to t+dvt+dv, where dd is the distance between the fronts of the two cells and vv is the velocity of the flowing cells.
We simulated the area-based dual-cell model, and then we studied the characteristics of the dual-cell model that corresponded to different distances between the two cells, as depicted in Figs. 3(a) and 3(c). To distinguish two cells in the fluorescent signals, a local minimum should exist between two adjacent local maxima, and the valley should be lower.33 Similar results could be observed using the volume-based dual-cell model, as shown in Figs. 3(b) and 3(d).
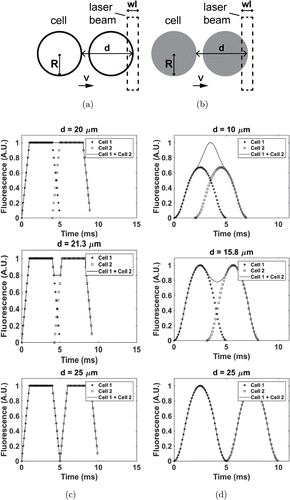
Fig. 3. Area-based and volume-based dual-cell models. (a) Phases of dual cells passing the laser beam in area-based model. (b) Phases of dual cells passing the laser beam in volume-based model. (c) Area-based dual-cell models with different cell distances. In this peak model, v=5v=5mm/s, k=1k=1mV/μμm2, R1=R2=10R1=R2=10μμm and W=5W=5μμm. (d) Volume-based dual-cell model with different cell distances. In this model, V=5V=5mm/s, k=1k=1mV/μμm2, R1=R2=10R1=R2=10μμm and W=5W=5μμm. A.U. denotes arbitrary unit.
3.4. Rayleigh criterion and algorithm to identify cell clusters
The resolution of in vivo flow cytometry was based on optical imaging. To distinguish two adjacent points, there should be some differences in the brightness of an overlapping area between the two diffraction spots. According to the Rayleigh criterion,34,35,36 the minimum value of brightness in the overlapping area should be 73.5% of the maximum value.
The algorithm to identify cell clusters was proposed based on the criterion. Briefly, the raw fluorescence data were denoised using a wavelet filter (Sym6, level 3). Then, the smoothed data trace was scanned to identify all local maxima, and a threshold was calculated according to the following equation :
As shown in Fig. 4(a), the effective signal for a cell was indicated by a circle. This group of signals contained only one single cell. In Fig. 4(b), there were two local maxima in this signal flock. However, the local minimum between these two local maxima was higher than 73.5% of the lower local maxima, indicated by the square. The lower peak was not regarded as an effective signal for the cell. Thus, one single cell was identified in this group of signals. A dual-cell cluster was identified, as shown in Fig. 4(c). The local minimum satisfied the Rayleigh criterion. A cell luster with four cells was identified in a signal group, as depicted in Fig. 4(d). This suggested that the proposed criterion and algorithm were effective in identifying cell cluster signals.
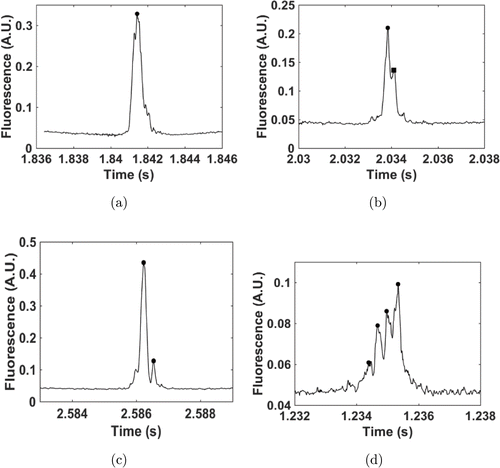
Fig. 4. Cell cluster identification using proposed algorithm based on Rayleigh criterion. (a) Typical data trace for single circulating tumor cell. Circle indicates position of local maximum. (b) Single cell signal identified by proposed algorithm. Square indicates local maximum, which is not considered an effective signal for cell. (c) Typical signal of cell cluster with dual cells. (d) Another typical signal of CTC cluster identified with proposed algorithm. This group of peaks indicates that this cell cluster consists of four cells. A.U. denotes arbitrary unit.
3.5. In vivo validation of proposed algorithm with intravital imaging
Intravital imaging was performed to validate the proposed algorithm. The GFP-labeled 4T1 breast cancer cells were injected into the blood of a mouse via the tail vein. Fluorescence images of blood vessels on the mouse ear were continuously captured in real-time. A representative image is shown in Fig. 5(a). Due to light absorption by hemoglobin, blood vessels appear dark in the image. The narrower vessel on the left is an artery, while the wider vessel on the right is a vein. A cell cluster consisting of two cells was observed in the vein. The part of the image within the dashed box was extracted to analyze the fluorescence profile. The fluorescence profile was calculated by integrating the fluorescence intensity within the black dotted box, which we then moved along the horizontal axis. The width of the box was ∼5∼5μμm, which was comparable to the laser slit in IVFC. Sliding the box mimicked a cell flowing through the laser slit. After getting the whole profile, it was normalized by the second largest profile value indicated by a black circle. The normalized valley profile value between the two peak profile values was below 0.735. This indicated that the group of signals represented a dual-cell cluster, complying with the Rayleigh criterion. An image of the cell cluster in the succeeding frame was shown in Fig. 5(c). Though intravital imaging was somewhat limited by motion blur, similar results were also observed. As shown in Fig. 5(d), the fluorescence profile of the cell cluster also complied with our proposed criterion.
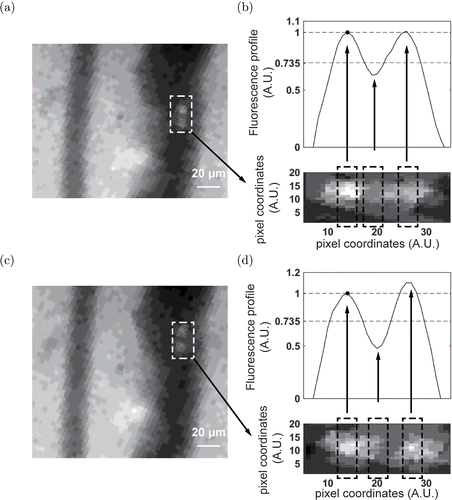
Fig. 5. In vivo validation of proposed algorithm with intravital fluorescence imaging. (a) Typical image of a 4T1 cell cluster with two GFP-labeled cells. (b) Normalized fluorescence profile of the cell cluster in (a). Fluorescence profiles were calculated by integrating the fluorescence intensity within the black dotted box, which slid along the horizontal axis. Width of box was ∼5∼5μμm, which was comparable to the laser slit in IVFC. Sliding the box mimicking the movement of a cell flowing through the laser slit. After obtaining the entire profile, it was normalized by the second largest profile value, indicated by a black circle. Normalized valley profile value between two peak profile values was below 0.735, indicating that the flock of signal represented a dual-cell cluster and complied with the Rayleigh criterion. (c) Image of cell cluster in succeeding frame. (d) Normalized fluorescence profile of cell cluster in (c). A.U. denotes arbitrary unit.
4. Discussion
In this work, we established two cell models for two different fluorescently labeled cells. The area-based model corresponds to cells with fluorescent molecules distributed on the surface of cell membrane, like DiD. The volume-based model can be applied to cells producing a fluorescent protein, such as GFP. Based on these two cell models, we analyzed dual-cell cluster signals, which represent the simplest type of cell clusters. Using the Rayleigh criterion, we developed a new algorithm to identify cell cluster signals. The effectiveness of the algorithm was verified with experiments.
A potential limitation of IVFC is that the collected signals provide no spatial resolution in the direction perpendicular to the blood flow. Two cells could pass through the laser slit side by side. The signals generated by these two patterns are barely distinguishable from single cell signals. In these situations, some cell clusters will be missed by IVFC. To obtain the spatial resolution, two possible solutions have been proposed and tested. One solution is to perform a line scan during data collection. The laser beam could be reshaped into a circle, and then the size of the laser beam could be reduced to generate a higher spatial resolution. A scanner could be used to move the laser spot along the direction perpendicular to the blood flow. The other solution is intravital imaging. By capturing images of flowing cells, single CTCs and cell clusters can be distinguished based on distinct morphological information. However, this solution also suffers from a limited frame rate.
Conflict of Interest
All authors declare no potential conflicts of interest.
Acknowledgments
This work was supported by a grant from the National Science Fund for Distinguished Young Scholars (Grant No. 61425006) and Program of Shanghai Technology Research Leader (Grant No. 17XD1402200).