Modulated illumination localization microscopy-enabled sub-10 nm resolution
Abstract
Optical microscopy is an essential tool for exploring the structures and activities of cells and tissues. To break the limit of resolution caused by diffraction, researchers have made continuous advances and innovations to improve the resolution of optical microscopy since the 1990s. These contributions, however, still make sub-10nm imaging an obstacle. Here, we name a series of technologies as modulated illumination localization microscopy (MILM), which makes ultra-high-resolution imaging practical. Besides, we review the recent progress since 2017 when MINFLUX was proposed and became the inspiration and foundation for the follow-up development of MILM. This review divides MILM into two types: point-scanning and wide-field. The schematics, principles and future research directions of MILM are discussed elaborately.
1. Introduction
From ancient times, people have begun the exploration of the microworld. As the most direct tool for observing the microworld, optical microscope has been widely used since its birth. Since the phenomenon of diffraction limit has been discovered by Abbe et al. in 1873,1 the resolution of conventional far-field microscopy has been constrained to diffraction limit, that is, , where NA is numerical aperture, and is wavelength. It was not until the second-half of the 20th century that the proposal and rapid development of fluorescence super-resolution microscopy made higher resolution possible.2 Moreover, two imaging modes, namely, point-scanning imaging and wide-field imaging, have created two development paths for optical far-field microscopy. Point-scanning microscopy illuminates a very tiny area with excitation light to emit fluorescence, then receives the fluorescence by unit detector and obtain global information by scanning through the whole field of view. The other mode is wide-field imaging, which relies on array detector to receive the fluorescence of the entire area emitted by wide-field illumination. In the follow-up research, people continue to improve the resolution of optical microscopes based on these two imaging modes, which provides technical support for studying the finer structures and more complex biological activities in cells and tissues.
Confocal microscopy, which appeared in the 1950s, uses a pinhole (PH) in the detection path of fluorescence microscope to produce a significant optical sectioning effect. It improved the resolution of the optical (fluorescence) microscopes by a factor of ,3 which greatly promotes the application of fluorescence microscopy in biomedical researches. Since the 1990s, super-resolution microscopy has reached its peak of development.4 Gustafsson.5 proposed structured illumination microscopy (SIM) based on wide-field microscopy in 2000, which utilizes specific modulation technology, such as interference, etc., to move high-order frequency information outside the optical transfer function (OTF) of the system into the OTF. These high-order frequency parts are not utilized in conventional microscopes. Limited by the influence of NA, the resolution of SIM cannot exceed twice the resolution of the original optical system, which constrains the spatial resolution of SIM to around 100–200nm.6 Although the two methods mentioned above break the diffraction limit and achieve better performance, due to the limit of NA and finite size of PH aperture, sub-100nm resolution is still hard to achieve, which cannot satisfy the ever-increasing biological application desire. The emergence of stimulated emission depletion microscopy (STED)7,8 and single molecule localization microscopy (SMLM),9,10,11 however, has changed this situation. With the help of continuous progress on fluorescent markers,12,13 STED and SMLM have successfully achieved imaging resolution with sub-100nm. Hell and Betzig, who proposed STED and photoactivated localization microscopy (a kind of SMLM), respectively, were also awarded the 2014 Nobel Prize in chemistry. STED was theoretically proposed in 19947 and implemented in 20008 by Hell et al. based on confocal microscopy. Since the focusing spot of excitation light is a spot with certain size but not an ideal point during the scanning process, the molecules within the spot will be excited and emit fluorescence to be collected by the detector. It is obvious that the resolution is influenced by the size of focal spot. Hell et al. used a beam of “doughnut”,14 which is also called STED doughnut, to nest around the excitation beam and match its wavelength with the wavelength of fluorescence. It makes the molecules excited to the excited state within the irradiation range of STED doughnut quickly fall back to the ground state without emitting fluorescence. Under the action of STED doughnut, only the molecules located at its center can emit fluorescence, which is equivalent to reducing the size of excitation spot and greatly improves the optical resolution. Theoretically, the resolution of STED has no limit. However, due to excitation light intensity, photobleaching and phototoxicity, its resolution is limited to 20–60nm. SMLM is a kind of wide-field microscopy, which controls the sparse luminescence of molecules in turn to determine the coordinate information of position by fitting the imaging spot of each molecule.15 The photons forming an imaging spot are equivalent to times of independent localization of the corresponding molecule. The theoretical localization precision is proportional to , where is the fluorescence central wavelength.16 If the excitation light is high enough and the imaging duration is long enough, the theoretical resolution of SMLM can also be infinitely small. Nonetheless, it is also limited by photobleaching and light-induced inactivation of the buffer, so its typical resolution is around 20–50nm, which is comparable to STED. Compared with methods such as SIM.17,18 and saturated SIM (SSIM),19,20 although SMLM and STED require higher-level sample preparation and slower imaging speed, they actually realize sub-100nm imaging. SIM does have a lower requirement for imaging conditions, but the frequency shift principle itself limits the further improvement of resolution. SSIM can achieve theoretically unlimited resolution, but its higher requirements for the excitation intensity and the “frequency-missing” phenomenon is easily generated at high-order frequency components, so it is not widely used.
It is worth mentioning that although the two types of methods, STED and SMLM, have achieved a real breakthrough in the resolution, their actual resolution is still limited by the imaging duration and illumination intensity, making it difficult for further improvement. Therefore, it is hard to meet the observation requirements of subcellular structures (below 10nm) only relying on STED and SMLM. Given this, in recent years, scholars have devoted themselves to developing ultra-high-resolution fluorescence microscopy. Minimal photon fluxes (MINFLUX), which combines modulated illumination with single molecule localization, was proposed by Hell et al. in 2017.21 MINFLUX uses doughnut-shaped illumination light to scan and excite single molecule in an area with a diameter of , collects the photons at four points around each molecule and combines the principle of triangulation to achieve single molecule localization. The theoretical localization precision is , where L is probing range and N is photon number. MINFLUX does not rely on extremely high intensity of excitation light. Instead, it requires the intensity to be as low as possible or even a doughnut with no brightness in the center. Based on this concept, the smaller the probing range, the higher the localization precision,21 but the slower the imaging speed. Because of the limited field of view and relatively long imaging duration of MINFLUX, it is generally used for imaging a small field of view or tracking and detecting a single molecule.22
To overcome the shortcomings of MINFLUX and retaining its ultra-high resolution, several single molecule localization wide-field microscopies based on modulated illumination have been proposed in recent years (Fig. 1), such as repetitive optical selective exposure (ROSE),23 SIMFLUX,24 structured illumination-based point localization estimator (SIMPLE)25 and modulation-enhanced localization microscopy (MELM)26 (C-MELM27). These methods also combine modulated illumination with SMLM and use the phase shift of illumination pattern to obtain corresponding distribution of photons of each molecule. With the help of photon number distribution and phase shift, the relative position between the single molecule and illumination pattern can be calculated. In the meantime, surpassing the localization precision of SMLM can be realized. In the case of same photon number, its resolution is about twice that of conventional SMLM. To some extent, it realizes the parallel processing of MINFLUX, which enables extended field of view and guarantees an identical precision for each molecule.
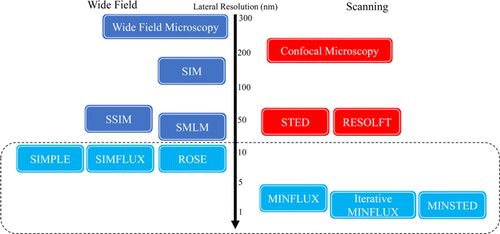
Fig. 1. (Color online) Comparison of main optical super-resolution microscopy methods. The performance comparison between the methods discussed in this paper and the current main super-resolution methods. The resolution performances shown in the figure are all lateral resolution. The light blue part is the main technology discussed in this paper, the red part contains point-scanning methods and the dark blue part contains the wide-field methods.
In addition to ultra-high resolution, observing thick samples is also essential for modern biomedical research, which places higher demands on three-dimensional (3D) detection capabilities. In general, the axial resolution of fluorescence microscope is significantly inferior to its lateral resolution. To achieve 3D detection, the structure of system is also much more complicated.28 Before 2014, 3D super-resolution microscopy was mainly achieved by improving two-dimensional (2D) microscopies such as SIM, STED and SMLM. In STED setup, 3D STED doughnut can be generated by adjusting spatial light modulator (SLM). And through spatial scanning, 3D detection can be realized. In SMLM, the axial resolution can be improved by introducing a cylindrical lens to introduce astigmatism in the axial direction29,30 or a 4Pi structure which consists of two opposing objectives.31,32 SIM and related methods can adjust the optical field distribution of structured illumination in the space so that the effective OTF is extended in the axial direction, which makes collecting high-order frequency information in axial direction possible.33,34,35 However, although these methods mentioned above improve the axial resolution, their 3D detection capabilities are not in an equivalent level. In most cases, the axial resolution is still not as good as lateral resolution, and there even exists a big gap.4 Since 2017, after the proposal of MINFLUX, combining single molecule localization and modulated illumination has become a new trend for 3D super-resolution microscopy. All methods, including point-scanning-based 3D MINFLUX,36 wide-field-based modulation localization (ModLoc)37 and ROSE-Z,38 improved the spatial resolution of fluorescence microscopy a lot (Fig. 2). These breakthroughs expand the application prospects of 3D super-resolution microscopy.
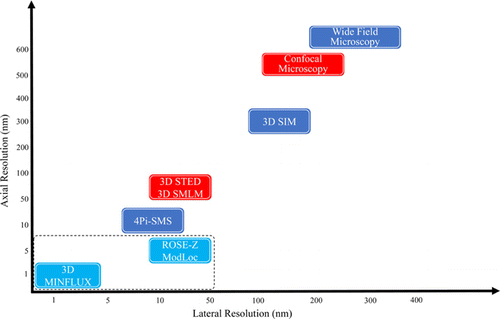
Fig. 2. (Color online) Comparison of main 3D optical super-resolution microscopy methods. The X-axis denotes the lateral resolution (localization precision); the Y-axis denotes the axial resolution (localization precision). The light blue part contains the main technologies discussed in this paper, the red part contains the point-scanning methods and the dark blue part contains the wide-field methods.
This review systematically introduces the technologies that combine single molecule localization and modulated illumination in the post-Nobel Prize era, whose resolution of three dimensions reaches 10nm or even less than 5nm. In the following chapters, this type of super-resolution microscopy is divided into point-scanning part and wide-field part, which can be named as modulated illumination localization microscopy (MILM), mainly including the principles, structures and applications.
2. Point-Scanning Imaging-Based MILM
2.1. Lateral super-resolution
The resolution of point-scanning fluorescence microscopy has been greatly improved because of STED, but technically, there are extremely high requirements for the STED doughnut. For example, only when the excited molecules return to the ground state by stimulated radiation whose rate is much higher than the rate of spontaneous emission emitting fluorescence, the STED doughnut can “erase” the fluorescence.3 Therefore, high-power pulsed LASER is required in the excitation process, and such LASER can easily cause irreversible damage to the sample. Consequently, achieving sub-10nm imaging is nearly impossible. Methods like ground state depletion (GSD)39 and reversible saturable optical fluorescence transition (RESOLFT)40 significantly reduced the high-power-density requirement of the STED doughnut by inhibiting the emission of part of the molecules.40 Although GSD and RESOLFT relax the requirement of hardware, the resolution and photon efficiency have not been obviously improved. In SMLM, achieving sub-10 nm resolution is also nearly impossible, that is, because collecting as many photons as possible to ensure the highest resolution extends the camera’s exposure time and imaging duration. Thus, the background noise and sample drift during the imaging process will become unignorable, and the credibility of SMLM is constrained.
To achieve 10nm or higher resolution, in 2017, with the help of the principle of SMLM, Hell et al. proposed MINFLUX which achieves localization precision of 3nm under the condition of collecting 500 photons21 (as shown in Figs. 3(a) and 3(b)). In the past super-resolution methods, such as STED and SMLM, it is necessary to detect and collect as many photons as possible in the process of image acquisition. Ideally, the stronger excitation light or the more acquired fluorescent photons, the better the resolution. However, MINFLUX is different from them. Specifically, the accurate position of a single molecule is determined by collecting only few fluorescent photons which follows the principle of Poisson distribution around the molecule. MINFLUX utilizes vortex phase plate (VPP) to form a doughnut as modulated excitation light and move the doughnut quickly in the focal plane to excite fluorescence. When the molecule is in the center of the doughnut, ideally, the avalanche photon diode (APD) will not collect any photon, and the photon count will be 015; when the center of doughnut is close to the single molecule, the closer it is, the fewer photons will be collected. Thus, the collected photons can not only affirm whether there are molecules near the position, but also reflect the relative position information between the single molecule and the doughnut center. Because of the special structure of doughnut, the signal which contains a small number of photons will be fully utilized to improve the resolution. In the meantime, another feature of MINFLUX is to correlate its localization precision with the probing range L, If the size of L is set to be extremely smaller than diffraction limit, the number of collected fluorescent photons will provide more information of the position of single molecule. To obtain accurate position of a single molecule, four detections are required in most cases. By placing the doughnut in four different positions around the initial guess of the single molecule, the number of fluorescence photons for each detection is obtained and the precise position of each molecule can be calculated. The four positions of detection are shown in Fig. 4(c), which are the position of initial guess and three vertices of the equilateral triangle centered on it.21 In 2020, Hell et al. used the influence of the probing range L on the localization precision to the extreme.36 Through iterations of localization and narrowing the probing range L in each iteration, the localization precision was further improved, which solved the problem of uneven precision in the range L by only one localization (four detections). It should be noted that although MINFLUX can achieve precise localization with a small number of photons, its localization still starts with the detection of the diffraction limit, and it is necessary to use traditional methods such as confocal to obtain the initial guess of each molecule. After obtaining the initial guess, the probing range L and the range of localization uncertainty will be reduced. What should be noted is that the essence of MINFLUX is still to maximize the acquisition of fluorescence detection information. The fluorescence efficiency is also much higher than that of traditional SMLM. Therefore, under the condition of same photon number, the resolution of MINFLUX is one order of magnitude than that of SMLM; and when the resolution is same, the number of photons used by MINFLUX is two orders of magnitude less than SMLM.41 As a point-scanning system, Hell et al. built MINFLUX system based on STED (shown in Fig. 3(b)).21 The STED doughnut is replaced by an excitation doughnut to achieve modulated illumination. To realize four rapid detection around each molecule, the entire system requires ultra-fast response and perfect coordination. Therefore, electro-optic modulator (EOM) is used to quickly to switch the ordinary focused beam and doughnut after passing through the dichroic mirror (DM), that is, to switch between confocal type (initial guess) and MINFLUX type (accurate localization).21 The rapid movement of the doughnut in the focal plane is executed by two orthogonal electro-optic deflectors (EOD) and a piezoelectric tilt mirror. The reason for using electro-optic devices is to accelerate the imaging speed and improve the live-tracking performance of MINFLUX. By expanding the LASER module, multicolor imaging and wide-field imaging can also be realized as an extension of MINFLUX.36
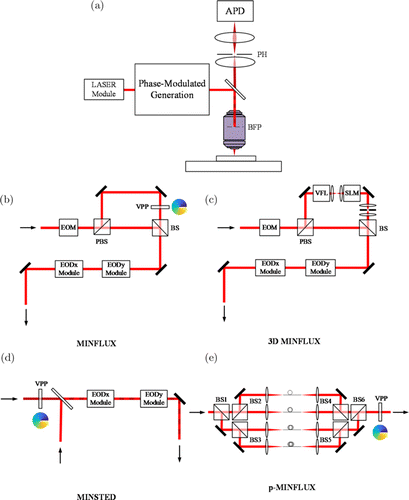
Fig. 3. Structure schematic diagram of point-scanning method of MILM. (a) Schematic diagram of the principle of the point-scanning system. The excitation light is concentrated on the focal plane of the sample. Emitted fluorescence passes through the deformable mirror and PH is finally received by the APD. The imaging of the entire field of view is realized by scanning. The main difference between the methods is the phase-modulated generation structure ((b)–(e)). (b) Modulated illumination structure of MINFLUX. (c) Modulated illumination structure of 3D MINFLUX. (d) Modulated illumination structure of MINSTED. (e) Modulated illumination structure of p-MINFLUX. EOM: electro-optic modulator, EOD: electro-optic deflector, BS: beam splitter, PBS: polarized beam splitter, VPP: vortex phase plate, VFL: electro-optically actuated varifocal lens, SLM: spatial light modulator, APD: avalanche photon diode, BFP: back focal plane, PH: pinhole.
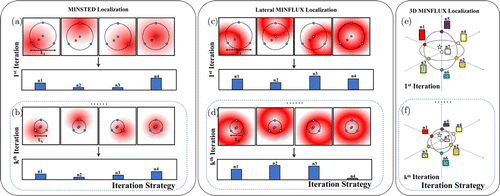
Fig. 4. Principle schematic diagram of point-scanning method of MILM. The star marks indicate the actual position of the single molecule, and the circular marks indicate the detection points. L1 represents the probing range of the first iteration, Lk represents the probing range of the kth iteration, and n1–n4 represent the number of fluorescent photons obtained by four detections. (a) MINSTED process, the scanning spot is E-PSF. (b) During the kth MINSTED iterative localization process, the probing range decreases as the number of iterations increases, and the size of the E-PSF also decreases. (c) MINFLUX process, the scanning spot is doughnut. (d) The kth MINFLUX iterative localization process, as the number of iterations of the detection range increases, the size of the doughnut does not change obviously. (e) 3D MINFLUX process, the scanning spot is 3D doughnut. The circular marks and histograms of different colors indicate the different detection points in one single localization and the number of fluorescent photons obtained, respectively. (f) The kth 3D MINFLUX iterative localization process.
However, the high hardware cost and difficulty of installation and adjustment of MINFLUX limit its application scale. In 2020, Tinnefeld et al., proposed p-MINFLUX42 which significantly reduce the hardware threshold of MINFLUX. They utilized pulsed interleaved LASER43 and beam splitters (BSs) in the system to obtain resolution of 1–3nm by only 1000–2000 photons. The normal MINFLUX needs pulsed LASER to form a doughnut and uses electro-optic devices to switch and move the doughnut, while using such devices must rely on field-programmable gate array (FPGA). Nevertheless, p-MINFLUX does not need to rely on FPGA and electro-optic devices. On the contrary, it divides the scanning beam into four paths through three BSs and enters four fibers with different lengths (as shown in Fig. 3(e)) and artificially generates time differences between each doughnut arrival at the focal plane to achieve rapid detection of each molecule.42 The four doughnuts focus on different positions around the molecule to complete a precise localization. Normal MINFLUX uses EOM and EOD to achieve fast scanning, and the time interval between each detection can reach 10–100s. By utilizing pulsed interleaved LASER and BSs, p-MINFLUX obtains an ultra-fast detection interval as low as 50ns which makes the time resolution of p-MINFLUX only dependent on the emission rate of fluorescence. Furthermore, p-MINFLUX is also able to obtain the fluorescence lifetime of every single molecule with the help of time correlated single photon counting to obtain the corresponding fluorescence lifetime imaging.44
The doughnut used in MINFLUX is inspired by STED doughnut, but it is the excitation light rather than the depletion light. Under ideal conditions, the intensity of the center of doughnut is 0 and the signal-to-background-ratio (SBR) will remain a high level.15 However, under actual circumstances, the excitation light intensity should be high enough to maximize the localization precision, which sacrifices the SBR. As the SBR decreases, the localization precision will drop accordingly. Aiming at alleviating this problem and improve the performance of MINFLUX, Hell et al. proposed Minimal Stimulated Emission Depletion (MINSTED) in 2020.45 Unlike MINFLUX which uses the center of doughnut for localization, MINSTED is closer to STED from the perspective of hardware [Fig. 3(d)]. It retains the 775nm depletion beam in STED and uses it to reduce the range of the 635nm Gaussian spot on the focal plane and form E-PSF, as shown in Fig. 4(a).45 Therefore, EOM that switches Gaussian excitation light and doughnut in MINFLUX setup can be removed. In short, in MINSTED, the doughnut used in MINFLUX is replaced by E-PSF46 which is formed by STED doughnut and excitation light. Since the depletion light and the excitation light share the same optical path and work together, MINSTED added an additional 633nm LASER to scan the entire field for the initial guess of each of molecule and provide priori information.45 MINSTED can achieve a resolution equivalent to that of MINFLUX but collect fewer photons. That is, because the STED doughnut not only reduces the range of Gaussian spot but also suppresses noise and artifacts. In this way, the problem that increasing the excitation light intensity reduces SBR is solved by MINSTED. Similarly, MINSTED does not use the peak of E-PSF to localize molecules, but rather uses the Gaussian tail to collect fluorescent photons (as shown in Figs. 4(a) and 4(b)), which possesses much higher photon efficiency.47 This conclusion is also proved in the wide-field counterparts.27
2.2. Axial super-resolution
As MINFLUX has brought lateral resolution of fluorescence microscopy to an unprecedented level, people have also begun to explore 3D imaging. In general, MINFLUX utilizes doughnut on the focal plane to localize the accurate position of molecules. This modulated spot, however, cannot improve the axial resolution. As a consequence, Hell et al. introduced 3D doughnut which is used in 3D STED48 into MINFLUX system and realize 3D imaging (unlike STED, both 2D and 3D doughnut are used as excitation light in MINFLUX instead of depletion light).36 To integrate 2D MINFLUX and 3D MINFLUX into just one system, as shown in Fig. 3(c), a SLM is introduced into the system to replace the VPP, and the corresponding 2D and 3D doughnut are generated by adjusting the SLM. Similarly, determining the accurate position of a single molecule needs multiple detections. After the initial guess by the confocal Gaussian spot, electro-optically actuated varifocal lens (VFL) is applied to localize the 3D doughnut above and below the molecule to obtain its initial axial position. If the priori information of spatial position is obtained, taking the initial guess as the center and choosing two symmetric detection positions about the center in -axis, -axis and -axis, respectively, to form a regular octahedron. Thus, seven detection positions besides the center point can be obtained (as shown in Fig. 4(e)). When the iterative algorithm is introduced, the probing range will decrease as the number of iterations increases, thereby improving the 3D resolution of 3D MINFLUX and the spatial uniformity of localization precision (as shown in Fig. 4(f)).49
Compared with conventional fluorescence microscopy, although MINFLUX improve both axial and lateral resolution by an order of magnitude, the complex system structure limits its promotion, especially for the more complex 3D MINFLUX itself — even the factors of vibration isolation and adaptation to the external environment will make MINFLUX imaging impractical.41 In 2021, Schmidt et al. developed a 3D MINFLUX system that can be used in a frame of ordinary microscope.41 This design builds a stabilizer based on the port of microscope stand and uses a closed-loop feedback mechanism in imaging process. The SLM can generate Gaussian spot, 2D doughnut and 3D doughnut, so EOM is unnecessary to be introduced into the system, which greatly simplifies the structure of excitation path.50 The deformable mirror is also applied in the system which can generate defocus phase difference to make the 3D doughnut move along the axial direction, and thus, the complicated VFL can be removed.41 3D MINFLUX and a series of revised systems makes imaging sub-cell structures and tracking single molecule with sub-5nm spatial resolution possible, which is revolutionary for biomedical researches. The fast-step improvement of 3D resolution has the potential for resolve and explain sub-cell activities and structures much more precisely. The ultra-high 3D resolution will also promote the development of related applications, like super-resolution correlative light and electron microscopy.
2.3. Conclusion
Point-scanning imaging-based MILM uses electro-optic devices such as EOM and EOD to perform ultra-fast switching and scanning the beams.21 By using SLM, deformable mirror and other adaptive optics devices, the excitation light can be modulated and moved rapidly.36 In the imaging process, this method estimates the initial position of each molecule by single molecule localization and confocal technology, and then generates modulated illumination to calculate the accurate position of each molecule based on fluorescence photon distribution and positional relationship. In this way, the photon efficiency can by maximized. Different from previous super-resolution methods, methods like MINFLUX, etc. no longer passively search the positions of molecules through the weak fluorescence on the detector, but actively move the center of doughnut or Gaussian tail as close as possible to the center of fluorescence. Consequently, MINFLUX can achieve the resolution of 1–3nm with collecting 1–2 orders of magnitude fewer fluorescence photons than conventional methods.36,41 Point-scanning imaging-based MILM, to some extent, are more suitable for single molecule tracking. That is, because imaging a large field of view requires to accurately localize numerous molecules.22 And it is relatively cumbersome and time-consuming to scan the whole field of view to obtain an image. In the follow-up research, if parallel detection and Airyscan51 can be combined to implement parallel MINFLUX, the imaging speed will be greatly improved and real-time tracking of multiple molecules under a large field of view may become practical.
3. Wide-Field Imaging-Based MILM
3.1. Lateral super-resolution
The main purpose of wide-field-based MILM, which is different from point-scanning-based counterpart, is to speed up the image acquisition process while maintaining ultra-high resolution. This type of microscopy is mainly based on SMLM, replacing the original wide-field uniform illumination with modulated illumination. As shown in Fig. 5(a), the localization precision is improved by using narrow-period fringes as an accurate “ruler”. The updated precise position coordinates of molecules are calculated by shifting the phase of the fringes (as shown in Fig. 6).
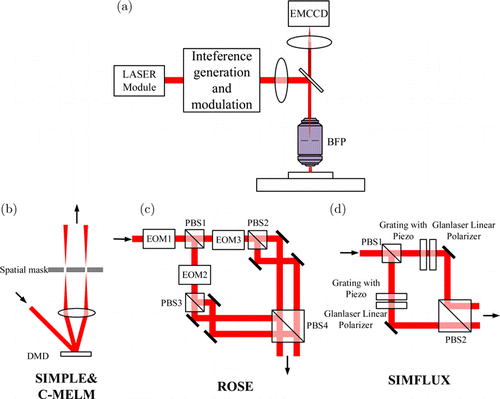
Fig. 5. Schematic diagram of the wide-field imaging-based MILM. (a) Schematic diagram of the principle of a wide-field microscope system. The excitation light is focused on the objective’s BFP after passing through the fringe generation and modulation system, and the objective illuminates the entire field of view. The fluorescence excited by the modulated fringes passes through the DM and converges on the array detector to achieve wide-field imaging. The main difference between methods is the composition of fringe generation and modulation system (b)–(d). (b) The modulated illumination structure of SIMPLE and C-MELM. (c) The modulated illumination structure of ROSE. (d) The modulated illumination structure of SIMFLUX.
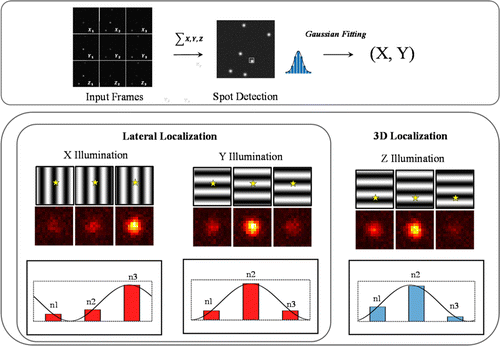
Fig. 6. Schematic diagram of the principle of wide-field imaging-based MILM. First, three images taken by a three-step phase shift of interference fringes in each direction are superimposed, and every single molecule is calculated by Gaussian fitting of SMLM to obtain the initial guess of the position. Then fluorescence intensity of every single molecule is extracted in the three-step phase shift of the interference fringe in different directions according to the initial guess. The phase in each direction is fitted to obtain the position of the single molecule relative to the interference fringe to improve the resolution.
SIMPLE is proposed by Reymond et al. in 2019,25 which applied digital micromirror device (DMD) to generate sinusoidal fringe and corresponding phase shift (as shown in Fig. 5(b)). By capturing and identifying photon number change of isolated emitters during the phase shift of sinusoidal fringes, SIMPLE can determine the relative position of each molecule to the fringes.25 Under the condition of low illumination intensity, the fluorescence intensity of a single molecule follows the linear response to excitation intensity. Therefore, for a single molecule, the parameters of the sinusoidal fringes can be obtained by fitting the three photon numbers collected by three-step phase shift and then, the relative position of single molecule in the fringes can be calculated. The fringes in the direction and direction can be used, respectively, to determine the corrected and coordinates of the single molecule. According to the experiments on single immobilized Alexa488 dyes, it is verified that comparing with SMLM, SIMPLE can improve the resolution by two times. It should be noted that SIMPLE has the advantages of high resolution and relatively low excitation intensity, making it possible for live cell imaging. In addition, it also puts forward the requirements on hardware: the imaging speed should be high enough to accomplish the collection of six images during the fringe direction switch and phase shifts actions in one on-state of a single molecule.52 Unluckily, the six images acquisition cycle in the process of SIMPLE, which contains more than one blinking period, will cause localization errors. In response to this issue, ROSE and SIMFLUX have solved it from a different perspective.
ROSE was proposed by Gu et al. in 2019.23 The system introduces three EOMs, in which the EOM in the main optical path works collaboratively with polarized BS (PBS) to switch two optical paths rapidly so that the high-speed switching of interference fringe direction can be realized. An EOM is set in each of the two optical paths to introduce phase difference to accomplish the phase shifts of fringe, as shown in Fig. 5(c). The excitation LASER is controlled by an acousto-optic tunable filter (AOTF), which reduces photobleaching and prolongs the fluorescence collection duration.23 The difference from other microscopies is that to solve the insufficient camera frame rate problem, ROSE applies a fast scanning galvanometer in the detection path and divides the six images mentioned above into six optical paths in sequence. Every three optical paths are collected by an electron-multiplying charge-coupled device. The acquisition of each image is only 125s (as shown in Fig. 7). The scanning galvanometer, AOTF, EOM and camera acquisition are controlled by high-precision synchronous clock signals to achieve image acquisition in ROSE. Compared with SIMPLE, ROSE introduces 405nm LASER for activating molecules and an LED system is applied to correct the drift of sample stage. ROSE uses DNA-orgami structures53,54 to make specific samples compare and analyze the localization precision between ROSE and ordinary SMLM which uses centroid fitting to localize molecules. It verifies that the resolution is improved by nearly two times which follows the results in simulations.23 In addition, Gu et al. also imaged the microtubule and filament structures of COS-7 cells, the results further verified that ROSE was able to improve resolution significantly.
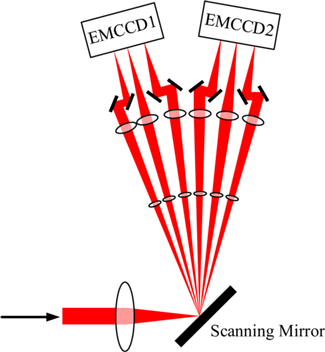
Fig. 7. Schematic diagram of fluorescence detection part of ROSE. The high-precision synchronization signal controls the LASER, camera and scanning mirror to work together, which can achieve a single frame acquisition speed of 125s when the camera exposure time is set for a relatively long duration, without introducing artifacts.
In 2020, Cnossen et al. proposed SIMFLUX,24 which uses Pockels cell and PBS to switch excitation light in two orthogonal directions rapidly and applied piezoelectric controlled grating to generate and shift the interference fringes in each direction, as shown in Fig. 5(d). Unlike ROSE, SIMFLUX made an algorithm to remove molecules that are not all at the same blinking period in the six images to ensure localization precision. Same as other methods, SIMFLUX sums up the six images and uses centroid fitting to obtain the initial guess of every molecule, and then calculate every molecule’s position relative to the fringe according to the intensities under different fringe phases to obtain the accurate coordinates. To make the fringe period more precise, SIMFLUX utilizes Fourier transform to estimate the fringe period in each direction.55 SIMFLUX also applied DNA-orgami structures to produce specific samples to compare its resolution improvement with ordinary SMLM. In addition, Cnossen et al. provided a detailed mathematical derivation for SIMFLUX,24 which provides principle support for wide-field-based MILM.
C-MELM was proposed by Schmidt et al. in 2021.27 It inherited the name from Reymond et al.,26 naming the wide-field MILM method as MELM. Schmidt et al. also emphasized that MELM is a kind of wide-field super-resolution method, so it is “camera-based”, that is, C-MELM.27 Current MELM can only use half of the localization information in images,56 so they proposed a MELM method based on illumination fringe priori, which enables to improve the resolution of current MELM as a factor of 2. For each molecule, PSF and image are both considered, and then the maximum likelihood estimation57,58 and a calibrated structured illumination model are used for position fitting. The hardware of C-MELM is like SIMPLE (as shown in Fig. 5(b)). The DMD and spatial filter are applied to generate interference fringe and accomplish phase shifts. The segmented polarizer27 is used to adjust the polarization of interference light to maximize the contrast of fringe.
3.2. Axial super-resolution
The concept of lateral MILM can also applied to axial localization. Similarly, the core idea is to determine the rough position of each emitter/molecule (the rough localization precision needs to reach the level of SMLM), and then calculate its relative position in the fringe. As shown in Fig. 6, the axial localization can still be improved by modulated illumination, but an initial guess before, that is, also essential, Which determines the final accurate localization precision.
In 2021, Jouchet et al. proposed a new method to improve the axial resolution and enables imaging of biological samples by up to several micrometers in depth, ModLoc,37 by introducing oblique interference fringes in the axial direction to achieve pure axial resolution improvement. Different from ROSE, SIMFLUX, etc., which rely on the fluorescence intensity fluctuation by phase shift to fit the relative position of each molecule in the fringe, ModLoc uses Pockels cell and PBS to introduce a fixed phase delay, obtaining four images with known phase delays, as shown in Fig. 8. The key parameters for calculating accurate axial position can be obtained through precise synchronized clock and mathematical derivations. In terms of optical design, ModLoc uses two beams to generate interference, one focusing on the center of back focal plane (BFP) of the objective and the other focusing on the edge of BFP, which generates oblique fringes in space. An EOM is set in one of the optical paths for phase shifting. In detection path, by combining BS and PBS, the detection path is divided into four paths. A quarter wave plate (QWP) and a Pockels cell are placed in each of path to introduce phase differences of , , , , respectively, which makes the four-channel fluorescence collected by detector meet the prerequisite of designed algorithm and calculate key parameters of axial position of each molecules.37 This method, however, is only used to improve axial resolution, and its axial resolution is limited by the localization precision of lateral direction. When the oblique fringe rotates to the limit angle, that is, entirely perpendicular to axial direction, it can be regard as lateral fringe and used for lateral localization. What is worthy of mentioning is that, due to its unique interference patterns, the approximate axial position of each molecule can be obtained when localizing the lateral position of the same molecule. Such a feature eliminates the influence of the axial period of the interference fringes when performing axial localization, which extends the imaging range of ModLoc to several micrometers, successfully realizing the observation and imaging of relatively thick samples.
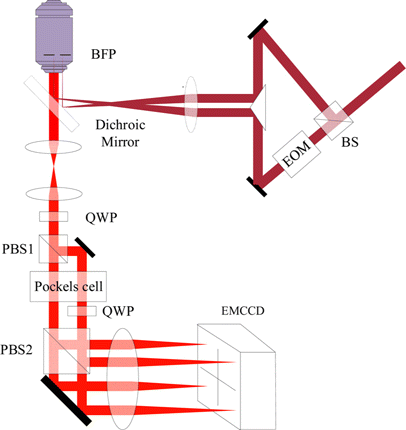
Fig. 8. Schematic diagram of ModLoc. Fluorescence excitation path: laser is divided by two beams by BS, one come through the EOM which is used to introduce a phase shift and the other did not, one focusing on the center of BFP and the other focusing on the edge. Fluorescence detection path: the fluorescence is divided into four channels, and the specified phase difference is introduced through the QWP and Pockels cell, and the high-precision synchronization signal works in synchronization with the laser. QWP: quarter wave plate.
In 2021, based on ROSE, Gu et al. proposed ROSE-Z38 and achieved single molecule localization with modulated illumination in axial direction by directly generate axial fringes. One beam of excitation light, which is originally incident on the BFP, is split to the other side of the sample by a BS and focuses on the sample. ROSE-Z uses parallel beam and convergent beam to generate interference fringes in the axial direction. And after three images are collected by three-step phase shift, the relative position of each molecule in the fringe is calculated using the same algorithm in ROSE. The difference between ROSE and ROSE-Z is that ROSE uses centroid fitting in SMLM to get the initial guess of lateral position, whereas ROSE-Z applies a cylindrical lens to generate astigmatism to obtain the initial guess of axial position. Compared with 4Pi systems of improving axial resolution like 4Pi-SMS,59 ROSE-Z only uses a single objective to receive fluorescence, which is easier to generate interference fringes and the system is more convenient to assemble.
3.3. Conclusion
The wide-field imaging-based MILM obtains the accurate position of each sparsely luminous single molecule in sequence and recovers a wide-field image by position superposition and relative algorithms. Compared with ordinary SMLM, this method uses modulated illumination to obtain multiple images with phase-shifted fringes and calculate the accurate position of each molecule through phase fitting. Due to the desire of completing switching fringe direction, phase shift and multiple shootings in just one on-state duration of a single molecule, the system requires extremely high synchronization accuracy and action speed.23,24,25,26 Therefore, the rapid switch and phase shift of fringes and the image recovery algorithm are the core of this technology. Unless introducing the fast scanning galvanometer like ROSE,23 the system requires very high camera frame rate. In addition, Image recovery algorithm is also essential. The quality of initial guess determines the precision of the entire algorithm,37,38 and phase fitting will also have a great impact on localization precision.60,61
Despite the high requirements for hardware and algorithm, this wide-field imaging-based MILM still reaches the best resolution among wide-field methods, which possesses essential significance for the observation of subcellular structures.
4. Summary and Outlook
The technologies we reviewed here are listed in Table 1. They are all fluorescence super-resolution microscopies that combine modulated illumination and single molecule localization, which have emerged since MINFLUX proposed in 2017. We divide them into two categories: point-scanning and wide-field, and discuss their principles, hardware and related applications. Whether it belongs to point-scanning method or wide-field method, their common superiority is ultra-high-resolution/localization precision. They all achieve sub-10nm, even sub-5nm resolution, which is nearly impossible to realize in former technologies, such as SIM, STED and SMLM. We can also find that the principle of these technologies has commonalities. They all originated from the understanding and extension of MINFLUX. Whether it is based on point-scanning system or on wide-field system, the essence is single molecule localization, which uses the sparse fluorescence emission of single molecule and the number of fluorophores to perform precise localization of molecules. The difference in focus is that technologies like MINFLUX increase the efficiency of fluorescent photons to an unprecedented level, thus, getting rid of the limitation on resolution caused by photobleaching and phototoxicity to some extent, which also makes ultra-high-resolution single molecule tracking in a small field of view possible. ROSE and similar technologies, however, focus on imaging with a large field of view. With the contributions of parallel modulated illumination, multiple molecules within the field of view can be accurately localized in the meantime, making ultra-high-resolution microscopy applicable by wide-field imaging.
| Method | Type | Lateral resolution | Axial resolution | Photobleaching/phototoxicity | Field of view |
|---|---|---|---|---|---|
| MINFLUX21 | Point-scanning | 1–3nm | — | Low | 10–15m |
| 3D MINFLUX36 | Point-scanning | 1–3nm | 1–3nm | Low | — |
| MINSTED45 | Point-scanning | 1–3nm | — | High | 10–15m |
| p-MINFLUX42 | Point-scanning | 1–2nm | — | Low | 10–15m |
| SIMPLE25 | Wide-field | 4.5nm | Same as SMLM | High | 20m |
| ROSE23 | Wide-field | 5nm | Same as SMLM | High | 25m |
| SIMFLUX24 | Wide-field | 8.6nm | Same as SMLM | High | 26m |
| ROSE-Z38 | Wide-field | Same as SMLM | 2nm | High | — |
| ModLoc37 | Wide-field | Same as SMLM | 6.8nm | High | — |
Point-scanning imaging-based MILM can achieve the highest resolution in fluorescence microscopy so far and increase the photon efficiency to an unprecedented level. At the same time, due to its full use of fluorescence photons, this technology also achieves ultra-high-resolution real-time single molecule tracking in a small field of view, which is evolutionary for studying complicated cell activities. Although MINFLUX is not the first one achieving single particle tracking,62,63 it carries forward the idea to an unprecedented level by combining fluorescence, modulated patterns and single molecule tracking. However, to realize ultra-high-resolution localization or tracking, technologies such as MINFLUX must face the rigorous hardware requirements. The application of SLM, EOM and EOD will make the imaging system complicated and unstable. Therefore, there are still obstacles for wide application of these technologies. Furthermore, this type of technology requires multiple detections of single molecule to achieve one localization and even requires iterative localizations for a single molecule. Its imaging speed, especially when imaging a large field of view, is limited. Thus, it is not that suitable for observing cells and tissues under a large field of view; wide-field imaging-based MILM is slightly inferior in resolution, but it still performs the best resolution among wide-field super-resolution microscopies at present. Besides, due to the fact that wide-field imaging-based MILM’s modulated illumination is closer to that of SIM, there are lower requirements for modulation hardware in the optical path than MINFLUX. Moreover, imaging with a large field of view can be achieved without the desire for sophisticated scanning devices. However, this kind of technologies are not perfect. Since all the steps, such as phase shifts, fringe switch and multiple image acquisition, should be finished in just one on-state of fluorescence, which implies higher requirements for the camera frame rate. In addition, because of the short fluorescence lifetime made by conventional fluorescent labeling methods such as small molecule probes and antibodies, which is usually only 10–20ms, it is still challenging for the camera to complete exposure and acquire image for many times in such a short time. Consequently, DNA-orgami is the commonly used sample structures, which limits the application scope.
With the emergency of technologies of MINFLUX, SIMFLUX, ROSE, SIMPLE and ModLoc, the imaging resolution has been continuously improved to a level of 10nm or less. Furthermore, the requirements for drift correction.64,65,66 and sample preparation strategy67,68 have become more stringent. The drift of hardware should be suppressed at a level much lower than the resolution of the system, which will be one of the most challengeable topics.21,41 It should be noted that the so-called fluorescence microscopy is the image of fluorescent dyes. How to use the position of the fluorescent dye to accurately represent the position of the observed target molecule is also a complicated interdisciplinary problem.
In the follow-up research for MILM, there is still room for optimization and innovation. For example, introducing parallel detection, Airyscan or 4Pi structure will further improve the performance of the system, but this put much higher requirements on the hardware and system assembling. Applying adaptive optics devices like deformable mirror can realize the correction of aberrations and the observation of thick samples, which has been verified in 4Pi system. Last but not the least, in fluorescence microscopy, high-quality super-resolution images are difficult to recover due to aberrations and noise. The application of algorithms such as deep learning69,70,71,72,73 and gradient descent74,75,76,77 can improve the quality of image post-processing and simplify the hardware to a certain extent. Especially for the technologies mentioned above, the rigorous requirements often limit the application scope and prospects.
The step of exploring the microworld will never be stopped. Luckily, optical super-resolution microscopy provides people with reliable tools. We anticipate that with the development of related techniques like sample preparation and image processing, the fluorescence microscopy based on single molecule localization combined with modulated illumination will offer more meaningful data and results for biomedical research in the future.
Conflicts of Interest
The authors declare that there are no conflicts of interest relevant to this paper.
Acknowledgments
This work was financially sponsored by National Natural Science Foundation of China (61735017, 61827825), Major Program of the Natural Science Foundation of Zhejiang Province (LD21F050002), Key Research and Development Program of Zhejiang Province (2020C01116), Fundamental Research Funds for the Central Universities (K20200132), Zhejiang Lab (2020MC0AE01) and Zhejiang Provincial Ten Thousand Plan for Young Top Talents (2020R52001). Y. S. and L. Y. contributed equally to this work.