GCR-Net: 3D Graph convolution-based residual network for robust reconstruction in cerenkov luminescence tomography
Abstract
Cerenkov Luminescence Tomography (CLT) is a novel and potential imaging modality which can display the three-dimensional distribution of radioactive probes. However, due to severe ill-posed inverse problem, obtaining accurate reconstruction results is still a challenge for traditional model-based methods. The recently emerged deep learning-based methods can directly learn the mapping relation between the surface photon intensity and the distribution of the radioactive source, which effectively improves the performance of CLT reconstruction. However, the previously proposed deep learning-based methods cannot work well when the order of input is disarranged. In this paper, a novel 3D graph convolution-based residual network, GCR-Net, is proposed, which can obtain a robust and accurate reconstruction result from the photon intensity of the surface. Additionally, it is proved that the network is insensitive to the order of input. The performance of this method was evaluated with numerical simulations and in vivo experiments. The results demonstrated that compared with the existing methods, the proposed method can achieve efficient and accurate reconstruction in localization and shape recovery by utilizing three-dimensional information.
1. Introduction
Cerenkov luminescence imaging (CLI) is an optical molecular imaging technology based on Cerenkov radiation, which is electromagnetic radiation emitted when a charged particle travels faster than light in the medium.1,2 Like other optical molecular imaging technologies, CLI has the essential advantage of high sensitivity and low cost. Beyond these advantages, it has wide availability in nuclide probes, which shows its great potential for clinical applications.3,4,5,6,7,8,9,10,11,12,13,14,15 At present, CLI has been used for tumor imaging, therapy monitoring, etc. However, CLI only performs planar imaging and cannot obtain the three-dimensional distribution of radionuclides, which hinders its further clinical application.
To trace the location and recover the distribution of radionuclides, the tomography method for CLI was proposed and named Cerenkov Luminescence Tomography (CLT).16 However, the reconstruction of CLT is a highly ill-posed reconstruction problem that makes it difficult to obtain accurate reconstruction results. To improve the reconstruction performance, various model-based methods were proposed for CLT as well as other optical molecular tomography technologies.17,18,19 Some researchers used prior information to improve the reconstruction accuracy.20,21,22,23,24 For example, Feng et al. took permissible region as prior information and proposed an optimal permissible region selection strategy.25 However, as the prior location of the source cannot be truly obtained, some researchers chose to reconstruct the internal source in the whole region.26,27,28,29,30 Zhong et al. proposed the whole body reconstruction algorithm along a regularization path.31 Besides the strategies mentioned above, taking multispectral information into account was also proposed.32,33 Spinelli et al. proposed a multispectral approach that used distinctive information content at different wavelengths in the reconstruction process, and the result demonstrated that multispectral information can improve the efficiency of CLT.34
The recent success in deep learning demonstrated its great potential in optical imaging tomography. By employing a deep neural network, the mapping relation between the surface photon intensity and internal radioactive source can be learned directly. Zhang et al. first applied a multilayer fully connected neural network (MFCNN) to reconstruct the distribution of the radioactive source.35 Cao et al. proposed a CLT reconstruction framework based on a stacked denoising auto-encoder, in which the residual error was learned between the preliminary reconstruction result and the accurate result.36 Zhang et al. applied an attention mechanism-based locally connected network for CLT, which improved the accuracy and stability of the CLT reconstruction.37 For the reconstruction in bioluminescence tomography and fluorescence molecular tomography, Meng et al. proposed a locally connected network based on the K-Nearest Neighbor method, which achieved accurate morphological FMT reconstruction in a short time.38 Yu et al. proposed a one-dimensional convolutional neural network, which significantly reduced the number of parameters.39 The methods mentioned above take the node intensity as the input of the network, and the nodes are generated by finite element method (FEM) software.20,26 However, the trained network is sensitive to the order of input, in other words, even for the same imaging model, if the order of input is disarranged, the trained network cannot work well.
In this paper, a 3D graph convolution-based neural network with a residual architecture for CLT reconstruction, named GCR-Net, is proposed to overcome the shortcomings of existing reconstruction methods based on deep learning. GCR-Net is divided into several residual blocks and each block consists of basic residual units. First, a three-dimensional graph convolution operator is introduced to extract and aggregate features. Second, coordinates and the surface photon intensity are performed as the input, which suggests that three-dimensional information is utilized in our network. It is noteworthy that the proposed network does not rely on a specific order for input points because of the characteristic of the three-dimensional graph convolution operator, which means that an accurate result can be obtained even if the order of input is disarranged.
The rest of this paper is structured as follows: In Sec. 2, basic knowledge of CLT reconstruction and the details of GCR-Net are introduced. In Sec. 3, the numerical simulation experiments as well as the in vivo studies are illustrated to prove the effectiveness of our network. In Sec. 4, the discussion and conclusion are illustrated.
2. Methodology
2.1. Model-based CLT reconstruction
For model-based CLT reconstruction, radiative transfer equation (RTE) is used to mathematically describe the propagation of light in biological tissue.40,41 However, RTE has high computational complexity. To simplify the equation, diffusion approximation (DA) model is proposed and widely used.16,42 Based on finite element analysis, a linear relation between the surface photon intensity and the distribution of the radioactive source is established, which can be defined as follows :
2.2. Deep learning-based CLT reconstruction
Deep learning-based CLT reconstruction uses neural network to establish the mapping relation between the photon intensity of surface and the radioactive source directly. Therefore, the deviation between the real photon propagation process and the simplified photon propagation process can be avoided. The deep learning-based method can be described as follows :
2.3. Three-dimensional graph convolution-based residual network for CLT
Generally speaking, the permissible domain can be discretized into nn nodes, which can be generated by a FEM software. Each node has three-dimensional information as well as photon intensity information. However, three-dimensional information has been ignored in the previous deep learning-based methods. Hence, a residual network based on 3D graph convolution for CLT, named GCR-Net, which fully utilizes 3D information is described in detail in this section.
2.3.1. Three-dimensional graph convolution
As each node and its neighbors can be organized as graph structure, and inspired by DGCNN,43 the EdgeConv layer is introduced into our network. In the EdgeConv layer, edge feature which describes the relationships between a node and its neighbors is proposed. It is defined as follows :
To update the features, a graph structure is in need. In our network, each node was treated as a vertex in the graph. The input graph was constructed by executing a K-NN search to find the nearest neighbor in 3D coordinate space, and the k was set to 20.
2.3.2. Network architecture
GCR-Net is based on residual architecture. The basic residual unit in our model is named GCUnit. GCUnit consists of three EdgeConv layers. Each EdgeConv layer is followed by a leaky-ReLU layer. A skip connection is added between the input and the output of GCUnit. As shown in Fig. 1, GCR-Net is composed of five residual blocks, named GCBlock. There are two types of GCBlock. One consists of two GCUnits and the other consists of three GCUnits. A skip connection is introduced in GCBlock to reuse a previously learned feature, which connects from the input of GCBlock to its output. Before entering the first GCBlock, an EdgeConv layer with eight output channels is performed, followed by a leaky-ReLU layer. At the end of the last GCBlock, an EdgeConv layer with 1 output channel is performed and then a leaky-ReLU layer is attached. The network configuration we used is shown in Table 1. Taking GCBlock 1 as an example, it consists of two GCUnits. Each GCUnit consists of three EdgeConv layers with 8, 16, and 16 output channels, respectively. It should be noted that, in our network, normalized coordinates and intensity are taken as inputs, and the size is n×4. The output of GCR-Net is the reconstructed distribution of the radioactive source.
| EdgeConv layer | GCBlock 1 | GCBlock 2 | GCBlock 3 | GCBlock 4 | GCBlock5 | EdgeConv layer |
|---|---|---|---|---|---|---|
| [8] | [81616]∗2 | [323264]∗3 | [6464128]∗2 | [643232]∗3 | [1688]∗2 | [1] |
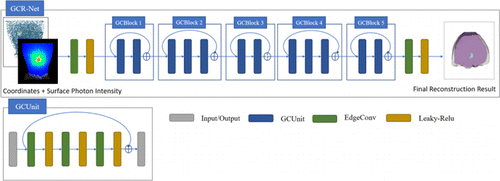
Fig. 1. The network structure of GCR-Net. Coordinates and surface photon intensity are taken as input, and the output is the final reconstruction result.
2.4. Implementation details and evaluation metrics
The training and test of GCR-Net were implemented using Pytorch and Python 3.7. All operation was performed on a personal computer with an AMD Ryzen 7 1700 Eight-Core Processor 3.00GHz CPU and a NVIDIA GeForce GTX 1080 Ti GPU. The optimization function of GCR-Net was Adam optimizer with a learning rate of 0.001. Mean Square Error (MSE) Loss was adopted as a loss function.
The performance of GCR-Net was quantitatively evaluated with the following three metrics: Location error (LE), Dice coefficient, and signal-to-noise ratio (SNR).
LE is defined as the position error between the reconstructed radioactive source and the actual radioactive source. Lr represents the barycenter of the reconstructed radioactive source and L0 represents the barycenter of the actual radioactive source. It can be defined as follows :
Dice coefficient is used to evaluate the similarity between the reconstructed source region and the actual source region. Srec represents the reconstructed source region and Sac represents the actual source region. It can be defined as follows :
The SNR is used to measure the visual quality. u′i represents the energy of the ith node in the reconstruction result, ui represents the energy of the ith node in the original numerical model. n is the total number of nodes in the numerical model. It can be defined as follows :
3. Experiments and Results
In this section, the performance of GCR-Net was evaluated by performing numerical simulations and in vivo experiments. IVTCG and MFCNN methods were used as baselines because they represent one of the most commonly used reconstruction methods in model-based methods and deep learning-based methods respectively.35,37 This section is structured as follows: The first part shows the reconstruction performance of our network in numerical simulation. It contains three experiments, including single-source experiments, dual-source experiments, and experiments of testing the influence of the order of input. The second part presents the reconstruction ability of our method in in vivo experiments.
3.1. Numerical simulation experiments
3.1.1. Data collection
A heterogeneous cylindrical phantom with a 10mm diameter and a 30mm height was designed to simulate the mouse body. The phantom consists of five kinds of organs: Heart, lungs, bone, liver, and muscle. The phantom is exhibited in Fig. 2. Optical parameters of all organs are presented in Table 2, which were obtained from the literature.44
| Component | Absorption coefficient μa(mm−1) | Scattering coefficient μs(mm−1) |
|---|---|---|
| Muscle | 0.016 | 0.510 |
| Heart | 0.011 | 1.053 |
| Lungs | 0.036 | 2.246 |
| Liver | 0.012 | 2.472 |
| Bone | 0.021 | 2.864 |
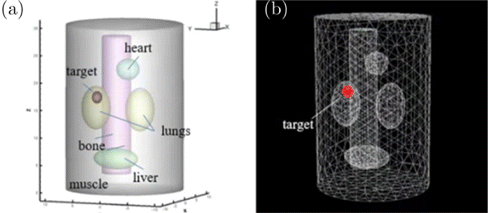
Fig. 2. The numerical phantom. (a) shows a single-source phantom, and (b) shows the standard mesh.
The numerical phantom was discretized into a standard mesh with 4626 nodes by using COMSOL Multiphysics V5.6 (COMSOL Inc., USA) software, from which its coordinates can be obtained.45 The order of input was determined at the discretized process by COMSOL Multiphysics. The standard mesh is exhibited in Fig. 2(b). A small spherical radioactive source with 1mm diameter was set in the model to present the tumor. The photon wavelength was set to 650nm to ensure appropriate penetration. As a data-driven method, plenty of data is in need for training. As it is difficult to collect a large amount of data from in vivo experiments, Monte Carlo method was used for data collection, because it can simulate the transmission process of photons. All the Monte Carlo simulation was conducted using Molecular Optical Simulation Environment (MOSE 2.3).46 We simulated a total of 221 single-source samples to make the radioactive source cover the permissible region as much as possible. 3000 dual-source samples were assembled by randomly selecting and adding the corresponding data of two single-source samples, which were as follows :
Overall, in this paper, 221 single-source samples and 3000 assembled dual-source samples have been simulated. 80% of the dataset was randomly selected for training and the others were selected for testing.
3.1.2. Single-source simulation result
To quantitatively analyze the effect of our network for single-source simulation, the mean value and deviation of different evaluation indexes were calculated, as exhibited in Table 3. As suggested from Table 3, GCR-Net obtained the lowest mean location error, highest mean Dice, and highest mean SNR. It demonstrated that the proposed network stably improved the accuracy in localization and morphology recovery. Furthermore, the network obtained the minimum LE value of 0.11mm and the maximum dice value of 0.79 in some cases, which further proved its advantages in terms of localization capability and morphology recovery ability.
| Method | LE (mm) | Dice | SNR |
|---|---|---|---|
| GCR-Net | 0.43±0.22 | 0.51±0.11 | 1.97±0.41 |
| MFCNN | 0.52±0.23 | 0.38±0.12 | 1.49±0.46 |
| IVTCG | 0.76±0.27 | 0.34±0.15 | 1.32±0.55 |
Two samples, named model 1 and model 2, were used to measure the single-source reconstruction result of different methods. In model 1, the source was set at (−3, −4, 3)mm, while the source in model 2 was set at (−6, −5, 17)mm. The quantitative results are exhibited in Fig. 3, which show that our network still performs better in LE, Dice, and SNR. Figure 4 shows the 3D visualization as well as the 2D cross sections of reconstruction results obtained by GCR-Net, MFCNN, and IVTCG, respectively.
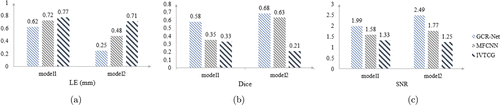
Fig. 3. The quantitative results of selected single-source samples. (a), (b), and (c) shows the quantitative value of LE, Dice, and SNR, respectively.
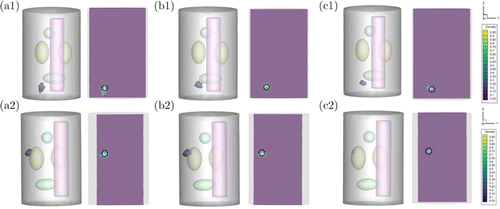
Fig. 4. Comparison results for the single-source experiments. (a1) presents the result reconstructed by GCR-Net method, (b1) and (c1) are the results obtained by the MFCNN method and IVTCG method in model1. (a2), (b2), and (c2) present the results reconstructed by GCR-Net, MFCNN, and IVTCG methods in model2, respectively. The black circle indicates the true position and size of the source.
Two additional single-source experiments have been conducted to prove our network is insensitive to the order of input. Two samples with single source, named model3 and model4, were used. In this experiment, the order of input has been disarranged. MFCNN was performed as a comparative experiment under the same conditions. It is noteworthy that the previously trained network was directly used without further training. The quantitative results are exhibited in Table 4. It can be demonstrated from Table 4 that MFCNN is sensitive to the input order. If the order of input is disarranged, the MFCNN cannot work well. On the contrary, GCR-Net can successfully reconstruct the distribution of the radioactive source with high accuracy in all experiments.
| Method | Model∖Metrics | LE (mm) | Dice | SNR |
|---|---|---|---|---|
| GCR-Net | Model 3 | 0.72 | 0.54 | 1.99 |
| Model 4 | 0.76 | 0.34 | 1.25 | |
| MFCNN | Model 3 | / | 0 | 0 |
| Model 4 | / | 0 | 0 |
3.1.3. Dual-source simulation result
Dual-source simulation experiments were carried out to further evaluate the performance of our methods on multi-source simulation. The quantitative results are exhibited in Table 5. The results in Table 5 revealed that our method still had its superiority in LE, Dice, and SNR, which presented a promising performance of reconstruction. However, compared with the single-source samples, the results of the dual-source samples were worse, which was probably caused by the raise in reconstruction complexity.
| Method | LE (mm) | Dice | SNR |
|---|---|---|---|
| GCR-Net | 0.69±0.25 | 0.37±0.10 | 1.68±0.45 |
| MFCNN | 0.75±0.28 | 0.31±0.11 | 1.29±0.47 |
| IVTCG | 0.91±0.33 | 0.16±0.11 | 1.04±0.50 |
Two samples, named model 5 and model 6, were used to measure the dual-source reconstruction result of different methods. In model 5, the centers of dual source were set at (−1, 3, 10)mm, (−3, −4, 22)mm, respectively. In model 6, the sources were set at (−2, −3.5, 4)mm, (−4.5, 0, 5)mm. The quantitative results are exhibited in Fig. 5, which illustrate a consistent trend with the former experiments. Figure 6 illustrates the visualization of the dual-source reconstruction results and suggests that both reconstructed radioactive sources have a clear shape and are easy to distinguish.
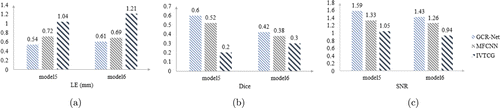
Fig. 5. The quantitative results of selected dual-source samples. (a)–(c) shows the quantitative value of LE, Dice, and SNR respectively.
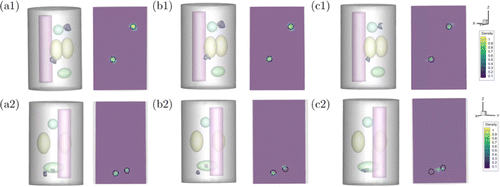
Fig. 6. Comparison results for the dual-source experiments. (a1) presents the result reconstructed by GCR-Net method, (b1) and (c1) are the results obtained by the MFCNN method and IVTCG method in model5. (a2), (b2), and (c2) presents the result reconstructed by GCR-Net, MFCNN, and IVTCG methods in model 6, respectively. The black circle indicates the true position and size of the source.
Two additional dual-source experiments have also been conducted to prove our network is insensitive to the order of input. Two samples with dual source, named model 7 and model 8, were used. The experimental setup was the same as the formal experiments on disarranging the order of input for single-source simulation. The quantitative results are shown in Table 6. It can be seen from Table 6 that in the dual-source experiment, when the order of input points changes, our network can achieve high accuracy in the reconstruction, while MFCNN still cannot work well.
| Method | Model∖Metrics | LE (mm) | Dice | SNR |
|---|---|---|---|---|
| GCR-Net | Model 7 | 0.56 | 0.52 | 1.83 |
| Model 8 | 0.58 | 0.37 | 1.29 | |
| MFCNN | Model 7 | / | 0 | 0 |
| Model 8 | / | 0 | 0 |
3.2. In vivo CLT reconstruction
The performance of our method was further evaluated with implanted small animal experimental data. The experimental data were from the CLT/micro-CT dual-modal system on adult nude mouse. The mouse was placed on the automatic rotating stage, which was rotating at 360 degrees with 1∘ interval to capture the images of CLI and X-ray projection. The CLI signal with wavelength of 650nm and white light data were collected using a cooled high-sensitivity EMCCD (−80∘, iXonEM Ultra 888, UK). The CT volume data was acquired by a micro-CT system (tube voltage of 40kV p, tube current of 300mA). The torso section of the mouse is shown in Fig. 7(a). We segmented the main organs of the mouse, including muscle, lung, heart, stomach, liver, and kidney, and integrated these organs into the mouse model. The optical parameters of each organ are consistent with those in the literature.47 A spherical glass vessel with a radius of 1mm containing about 800±50μCi 18F-FDG was used to mimic the lesion containing a radionuclide probe, which was implanted in the designed location. Animal care and protocols were approved by the Fourth Military Medical University Animal Studies Committee. The mouse was used twice for the single-source experiment and dual-source experiment, respectively. The position of the source was set at a different place each time. In single-source experiments, the source was implanted into the mouse at a specific location (22, 20, 31.5)mm. In dual-source experiments, the sources were implanted at (20.5, 27.5, 16.5)mm, (14.5, 20, 36)mm, respectively. The CLI imaging obtained from each implanted model was mapped and registered to the surface of the mouse torso. GCR-Net, MFCNN, and IVTCG methods were used to carry out the single-source experiment and dual-source experiment. The results are shown in Table 7 and Fig. 7, which prove the superiority of our network in CLT reconstruction for in vivo experiments.
| Single-source experiment | Dual-source experiment | |||||
|---|---|---|---|---|---|---|
| Method | LE (mm) | Dice | SNR | LE (mm) | Dice | SNR |
| GCR-Net | 0.37 | 0.81 | 1.76 | 0.54 | 0.87 | 1.89 |
| MFCNN | 0.47 | 0.66 | 1.23 | 0.72 | 0.69 | 1.46 |
| IVTCG | 0.48 | 0.65 | 1.14 | 0.96 | 0.50 | 1.01 |
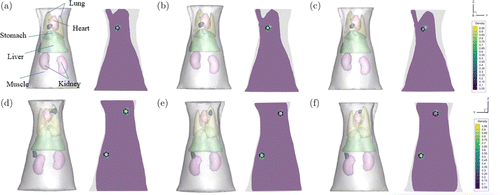
Fig. 7. Comparison results for in vivo experiments. (a) presents the torso section and the result reconstructed by GCR-Net method, (b) and (c) are the results obtained by the MFCNN method and IVTCG method for the single-source experiment. (d)–(f) presents the results reconstructed by GCR-Net, MFCNN, and IVTCG methods for the dual-source experiment. The black circle indicates the true position and size of the source.
4. Discussion and Conclusion
In this paper, a novel GCR-Net was proposed to achieve the accurate reconstruction of CLT. GCR-Net was built by stacking residual blocks based on 3D graph convolution. The main advantages of our network are summarized as follows: First, our network fully utilizes three-dimensional information and improves the performance of CLT reconstruction. Secondly, our network is insensitive to the order of input, which is not able to be satisfied by the traditional fully connected network.
To verify the performance of GCR-Net, single-source and dual-source numerical simulations were implemented. IVTCG method and MFCNN method were adopted for comparison. Quantitative analysis demonstrated that the GCR-Net showed a better performance in source localization and morphology recovery. To further evaluate the practicability of GCR-Net, in vivo experiments were performed. Consistent with our numerical simulation experiments, GCR-Net obtained the most accurate reconstruction results of radioactive probe distribution in biological tissue.
It is worth noting that our network is insensitive to the order of input. Our method can achieve accurate reconstruction results even if the order is disarranged. It can be explained by the characteristic of the EdgeConv layer. The EdgeConv layer is designed to be invariant to the ordering of neighbors. In EdgeConv layer, the applied aggregation function is a symmetric function. Thus, the output of EdgeConv layer is invariant to the permutation of the input.
However, there are still some limitations of GCR-Net. When calculating the edge feature set, the memory cost will be significantly increased with a higher setting of k in KNN method. Besides, the utilization of standard mesh depends on certain software and the registration between standard mesh and actual structure probably brings extra error in CLT reconstruction.
In conclusion, GCR-Net is proposed for CLT reconstruction. It is based on the 3D graph convolution and has the advantage of being insensitive to the order of input, which can achieve accurate reconstruction in source localization and morphology recovery. We hope our network can assist CLT reconstruction efficiently.
Conflicts of Interest
The authors declare that there are no conflicts of interest related to this article.
Acknowledgments
National Key Research and Development Program of China (2019YFC1521102); National Natural Science Foundation of China (61701403, 61806164, 62101439, 61906154); China Postdoctoral Science Foundation (2018M643719); Natural Science Foundation of Shaanxi Province (2020JQ-601); Young Talent Support Program of the Shaanxi Association for Science and Technology (20190107); Key Research and Development Program of Shaanxi Province (2019GY-215, 2021ZDLSF06-04); Major research and development project of Qinghai (2020-SF-143).


