SpectraTr: A novel deep learning model for qualitative analysis of drug spectroscopy based on transformer structure
Abstract
The drug supervision methods based on near-infrared spectroscopy analysis are heavily dependent on the chemometrics model which characterizes the relationship between spectral data and drug categories. The preliminary application of convolution neural network in spectral analysis demonstrates excellent end-to-end prediction ability, but it is sensitive to the hyper-parameters of the network. The transformer is a deep-learning model based on self-attention mechanism that compares convolutional neural networks (CNNs) in predictive performance and has an easy-to-design model structure. Hence, a novel calibration model named SpectraTr, based on the transformer structure, is proposed and used for the qualitative analysis of drug spectrum. The experimental results of seven classes of drug and 18 classes of drug show that the proposed SpectraTr model can automatically extract features from a huge number of spectra, is not dependent on pre-processing algorithms, and is insensitive to model hyperparameters. When the ratio of the training set to test set is 8:2, the prediction accuracy of the SpectraTr model reaches 100% and 99.52%, respectively, which outperforms PLS_DA, SVM, SAE, and CNN. The model is also tested on a public drug data set, and achieved classification accuracy of 96.97% without pre-processing algorithm, which is 34.85%, 28.28%, 5.05%, and 2.73% higher than PLS_DA, SVM, SAE, and CNN, respectively. The research shows that the SpectraTr model performs exceptionally well in spectral analysis and is expected to be a novel deep calibration model after Autoencoder networks (AEs) and CNN.
1. Introduction
The active ingredients of the same type of drug produced by different manufacturers differ due to distinctions in raw materials, manufacturing processes, resulting in price differences. Unscrupulous merchants take the similarity of active ingredients in drugs to package low-cost drugs into high-priced drugs from well-known manufacturers. This happens from time to time, seriously damaging consumers’ rights and interests as well as the reputations of well-known manufacturers. Therefore, it is critical to identify drug manufacturers during drug supervision.
Traditional methods for the identification of drug manufacturers include chemical methods, HPLC authorization methods, etc. But the detection process is cumbersome, the analysis speed is slow and destructive. As one of the mainstream nondestructive testing techniques, near-infrared spectroscopy has been widely used in many fields due to its fast detection speed and low cost.1,2,3,4 However, the drug supervision methods based on spectral analysis are heavily dependent on the chemometrics model which characterizes the relationship between spectral data and drug categories. The resolution of spectroscopic measuring devices has grown in tandem with the advancement of manufacturing technology. While more information about the analysis target can be obtained, the high-dimensional and easily disturbed spectra also make it difficult for traditional chemometrics to directly extract effective features. Therefore, pre-processing and wavelength selection algorithms are needed prior to modeling.5,6,7 At the same time, due to the wide variety of existing drugs and the continuous research and development of new drugs, the cumulative drug data also poses challenges to methods such as PLS and SVM.8
The application of advanced methods represented by deep learning in spectroscopy analysis promotes the development of chemometrics. Autoencoder networks (AEs) and convolutional neural networks (CNNs) are the most common deep learning-based spectral analysis approaches currently available. The former uses an autoencoder network to reconstruct features and then classifies them with traditional classifiers, while the latter extracts feature based on convolutional layers and use fully connected layers for classification.9,10,11,12 Although both can automatically learn and extract features from a large volume of spectral data, with low dependency on preprocessing and wavelength selection engineering, some studies have shown that convolution neural networks perform better in applications.13,14,15 However, CNNs have many hyperparameters, and their prediction performance depends on appropriate hyperparameter selection, which leads to difficult network construction.16,17,18,19 According to the most recent deep learning research, the transformer model based on the attention mechanism has the feature extraction potential of the CNN model and is easier to build network parameters. It is one of the current research hotspots of deep learning.20,21,22 Some studies have applied transformers to various fields such as drug design,23 chemical synthesis,24 protein three-dimensional structure prediction,25 medical imaging,26 and hyperspectral analysis,27 and demonstrated excellent results. But its application potential in molecular spectroscopy has not been studied yet.
This paper proposes a novel deep learning model for drug spectral identification, in order to build an analytical model with the better predictive ability and lower hyperparameter dependence. The approach is named SpectraTr based on the transformer structure. The performance of the model was verified based on the spectral data sets of our drugs from multiple manufacturers, and then the public drug data sets were used to further prove the advanced nature of the model. In the following paper, we will introduce our experiment materials and the mechanism of the transformer network structure in Sec. 2. In Sec. 3, the experiment procedure and the classification prediction results in different datasets will be discussed. Section 4 will summarize the whole paper. The source code is available at https://github.com/FuSiry/Transformer-for-Nirs for academic use only.
2. Materials and Methods
2.1. Datasets
Dataset A is provided by the National Institute for Food and Drug Control and all samples pass the statutory method test. It comes from 333 cefixime drugs produced by Guangzhou Baiyunshan Pharmaceutical General Factory and 716 phenytoin sodium drugs produced by 11 pharmaceutical companies including Southwest Pharmaceutical Co., Ltd. The raw spectra of both drugs are shown in Fig. 1 and are measured by the Bruker Matrix spectrometer. Each spectral wavelength ranges from 4000 to 11995cm−1−1, with intervals of 4cm−1−1 and a total of 2074 absorption points. It can be seen from the figure that the spectra of similar drugs from different manufacturers are similar.
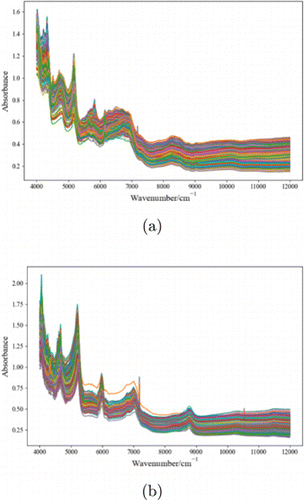
Fig. 1. Spectra of cefixime and phenytoin sodium tablets. (a) Spectra of cefixime tablets produced by seven pharmaceutical companies and (b) Spectra of phenytoin sodium tablets produced by 11 companies.
Dataset B is an open-source drug tablet dataset with a wavelength range of 7400–10507cm−1−1 and 404 absorbances points. The samples were classified into four categories according to their active ingredients. The raw spectra are shown in Fig. 2 and can be obtained from http://www.models.life.ku.dk/Tablets.
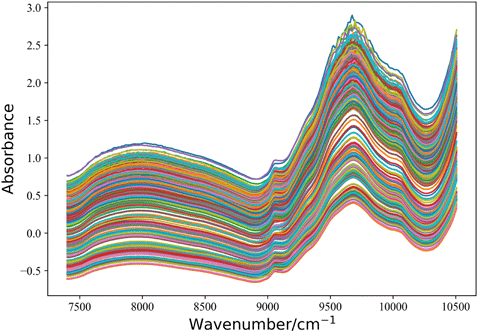
Fig. 2. Spectrum of drug table dataset.
2.2. SpectraTr architecture
As one of the most groundbreaking works in the deep learning field, the transformer has been widely discussed. We propose SpectraTr, as shown in Fig. 3, based on its work.28 SpectraTr is similar to a classifier based on AE network and consists of an encoder and MLP layers. The raw spectrum is divided into several spectra patches. After the position coding, the spectral features are extracted from the Spectra encoder composed of multi-head attention layers and weighted by MLP layer, which obtains the qualitative analysis results of the drug spectrum.
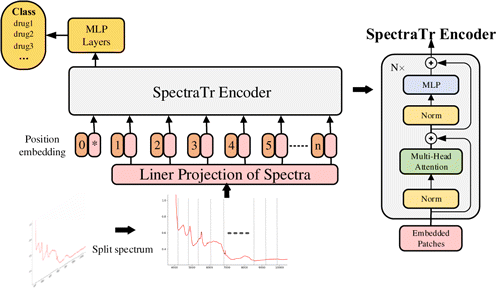
Fig. 3. The architecture of SpectraTr.
2.2.1. Position embedding
SpectraTr splits the full spectrum into multiple spectra patch sequences and extracts the features, which makes it compatible with 1d spectrum and 2d image data. Previous researchers have made similar attempts in the CNN network by splitting the raw spectrum into multiple segments and then entering the model, with better results.16 However, the position information of the spectra patch in the raw spectrum is destroyed because of the splitting of the spectrum into multi-segment spectra patches. In order to retain the position information, the position encoding is performed on the spectra patch, as shown in the following equation :
2.2.2. Multi-head attention mechanism
The self-attention mechanism is the core innovation of the transformer model. It can automatically focus on the important information in the data, increase the weight of important information and decrease the weight of unimportant information in the input data. The implementation of the attention mechanism in chemometrics is similar to automatic wavelength selection.29 The computational procedure of the self-attention mechanism is shown in Fig. 4.
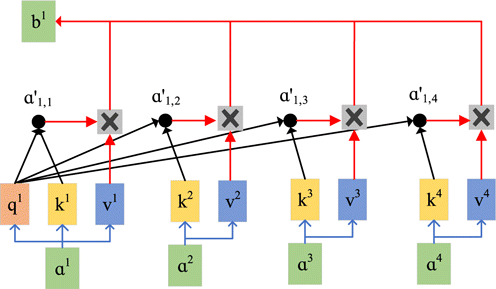
Fig. 4. The computational procedure of the self-attention mechanism.
The matrices qiqi, kiki, and vivi are obtained by linear transforming the input sequence aiai. Then, the elements of matrix qiqi from aiai are dot-product with the corresponding elements of the matrix kjkj from ajaj to obtain aijaij, which is the correlation between aiai and ajaj, as shown in the following equation :
Finally, the relationship degree of each input vector with other vectors a′i,j is multiplied by vj, to obtain the attention score of the input, as shown in Eq. (4). If ai is strongly correlated with aj, the calculated value of a′i,j will be larger. Eventually, oi,j will be higher, representing the information is more critical.
Multiple self-attention layers are used to form a multi-head attention block to capture richer feature information. This is similar to the principle of using multiple filters simultaneously in CNN. The schematic is shown in Fig. 5.
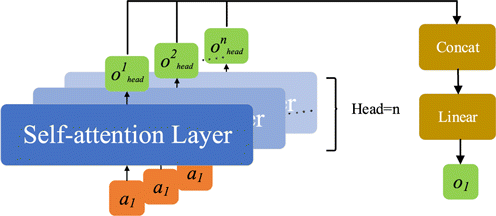
Fig. 5. Structure diagram of multi-head attention mechanism.
A single input a1 is sequentially stacked through the output matrix of n self-attention layer. Then, the result is linear matrix transformed to obtain the final attention matrix o1. Multiple self-attention layers form multiple subspaces, which allow the model to focus on different aspects of information.
2.2.3. Layersnorm and residual connection
A layersnorm (LN layer) is added before and after the multi-head attention block to eliminate the undesirable effects caused by odd sample data, which is similar to the principle of using Batchnorm (BN) in CNN, as shown in the following equation :
Since the spectral encoder also uses a stacked structure similar to CNN, an over-deep network structure may cause the gradient disappearance problem. Therefore, drawing on the idea of Resnet, a residual mechanism is introduced between the multi-head attention block and the MLP block, as shown in the following equation30 :
2.3. Modeling
SpectraTr is implemented based on Python 3.8 and the open-source deep learning framework Pytorch 1.8.0. It runs on workstations equipped with GeForce MP40GPU, Intel®Corei7 4770K CPU @ 3.5GHz and ubuntu operating system. The network structure of SpectraTr is described in Sec. 2.2. The initial parameters are shown in Table 1.
| Hyperparameter | Type | Range |
|---|---|---|
| Batch size | MT | {16, 32, 64, 128} |
| Learning rate | MT | {1e-4–1e-1} |
| L2 regularization | MT | {0–3e-2} |
| Patch num | MA | {5, 10, 20, 25, 40, 50, 80, 100} |
| Multi-head num | MA | {8, 10, 12, 14, 16, 18, 20} |
| Layer depth | MA | {2, 3, 4, 5, 6, 7, 8, 9} |
| Mlp_dim | MA | {128, 256, 512, 1024, 1536, 2048} |
On both datasets, cross-entropy is used as the loss function and the Adam optimizer is used for training. The total epoch is set to 200. In order to balance the training time and training effect, the learning rate decline strategy is introduced. If the loss of the training set does not decline within 10 epochs, the learning rate becomes half of the current one. The early_stop strategy is also introduced to prevent over-fitting. If the loss of the test set does not drop within 30 epochs, the training stops. The final selected parameters are shown in Table 2.
| Hyperparameter | |||||||
|---|---|---|---|---|---|---|---|
| Dataset | Batch size | Learning rate | L2 regularization | Patch num | Multi-head num | Layer depth | Mlp_dim |
| A | 16 | 0.0001 | 0.01 | 40 | 19 | 5 | 512 |
| B | 16 | 0.0001 | 0 | 10 | 12 | 3 | 1024 |
2.4. Baseline comparison
The classical chemometrics model SVM, PLS_DA, and the mainstream deep learning model SAE and CNN network are used as baseline models, respectively and compared with the proposed SpectraTr. SVM and PLS_DA are implemented by the machine learning library scikit-learn, where SVM parameter C is set to 1.0, gamma is set to 0.001, PLS_DA component is set to 22, and the rest is the default. The SAE network consists of three encoder stacks of input-1024, 1024-256, 256-output, as in the reference study.8 The CNN network consists of three convolution layers and three fully connected layers, as in the reference study.13 The above model is the same as the operating environment for SpectraTr.
2.5. Model evaluation
Models are evaluated using the classical qualitative analysis metrics: accuracy, specificity, recall, and F1-Score. Accuracy is the number of samples predicted correctly as a percentage of the total. Precision represents the percentage of positive samples predicted by the model that is actually positive. The recall represents the percentage of samples actually positive that are predicted to be positive. F1-score is the result of reconciling precision and recall. Their mathematical expressions are shown in the following equation :
3. Result and Discussion
3.1. Result of dataset A
3.1.1. Performance of spetralTr in dataset A
In this section, based on the proposed SpectraTr model, the near-infrared spectroscopy of cefixime and phenytoin sodium produced by multiple manufacturers are predicted and compared with other baseline methods. For comparison with previous research, we use the same dataset as in Ref. 13 and follow the arrangement of experiments described in the text. First, a qualitative analysis of cefixime tablets from seven manufacturers was carried out. Then, the mixture of cefixime tablets and phenytoin sodium tablets was used for qualitative analysis of 18 manufacturers in two drug classes. In experiments, 80% of the samples were randomly selected as the train set, and the remaining 20% were used as the test set. Both train and test sets use standardized pre-processing to speed up the model training, and all experimental results come from the test set. The prediction performance of SpectraTr on cefixime and mixed drugs is shown in Table 3.
| Dataset | Acc | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Cefixime | 100.00 | 100.00 | 100.00 | 100.00 |
| Phenytoin sodium and Cefixime | 99.52 | 99.55 | 99.52 | 99.53 |
The proposed SpectraTr model has excellent manufacturer identification capabilities for both cefixime and mixed drug spectra. The prediction accuracy on the cefixime data set of seven manufacturers achieves 100%, and prediction accuracy on the spectrum data set of 18 manufacturers of mixed drugs also achieves 99.5%.
3.1.2. Model performance under different training sets
The usual spectrum data is less due to the expensive cost of gathering spectral data and labeling it. However, deep learning is driven by the data. It is easy to overfit and leads to poor prediction performance with fewer samples. Therefore, the training set is randomly selected from the drug spectroscopy data set at a ratio of 0.8 to 0.2 to construct training sets and testing at different scales. Comparing SpectraTr with the baseline model, the accuracy is taken as the evaluation index, and the result on cefixime drugs is shown in Table 4.
| Train/Test | SVM | PLS_DA | SAE | CNN | SpectraTr |
|---|---|---|---|---|---|
| 266/67 | 97.61 | 96.05 | 99.01 | 100.00 | 100.00 |
| 233/100 | 97.00 | 95.00 | 99.00 | 100.00 | 100.00 |
| 199/134 | 96.25 | 94.06 | 98.76 | 99.25 | 99.83 |
| 166/167 | 95.20 | 93.40 | 98.28 | 98.70 | 99.57 |
| 133/200 | 94.00 | 91.37 | 97.80 | 98.23 | 99.04 |
| 99/234 | 92.87 | 89.15 | 96.86 | 97.50 | 98.58 |
| 66/267 | 90.63 | 87.21 | 95.32 | 96.38 | 97.06 |
The prediction accuracy of the deep learning model is better than that of the classic chemometric method when the pre-processing method is not precisely chosen, and there is no notable performance erosion when the sample size is reduced. The proposed SpectraTr model performs best on every different scale train set. The prediction accuracy even reaches 100% when the train set is 266 and 233. After the train set is reduced to 66, the accuracy is still 97.06%, which is slightly better than CNN and SAE, and significantly better than PLS_DA and SVM. Construct train sets with various proportions and conduct studies on mixed drugs in the same way. The result is shown in Table 5.
| Train/Test | SVM | PLS_DA | SAE | CNN | SpectraTr |
|---|---|---|---|---|---|
| 839/210 | 88.57 | 92.00 | 99.49 | 99.57 | 99.52 |
| 734/315 | 85.39 | 91.36 | 99.28 | 99.64 | 99.68 |
| 629/420 | 84.04 | 89.76 | 98.09 | 98.86 | 99.07 |
| 524/525 | 81.90 | 87.90 | 96.81 | 98.01 | 98.67 |
| 419/630 | 78.10 | 85.13 | 95.77 | 96.90 | 97.65 |
| 314/735 | 77.14 | 82.59 | 94.73 | 94.16 | 95.98 |
| 209/840 | 74.40 | 79.28 | 92.36 | 92.75 | 93.81 |
The results of deep learning methods are still better, similar to the results on cefixime. In most cases, SpectraTr is the best model in the majority of scenarios. It achieves a prediction accuracy of 99.68% when the training sample is 734. When the training sample is reduced to 209, the result is still 93.81%, which is better than CNN and SAE. However, when the train set is reduced, the performance loss is more noticeable than on cefixime, indicating that deep learning performance in small samples is also restricted by the data set’s classification difficulty.
3.1.3. Influence of pre-processing algorithm on model prediction ability
The drug is pre-processed with the methods of standardization, MSC, SG (window is 15), and SNV, respectively, to compare the effects of different pre-processing algorithms on the model. Under the 7:3 train set and test set division, the experimental results of the model on the data set A are shown in Table 6. SVM, PLS_DA are greatly affected by the pre-processing algorithm, under the suitable pre-processing can have more than 90% accuracy rates. But in most cases, the model does not have usability. Deep learning shows a low dependence on pre-processing. Although different pre-processing methods have different predictive performances, they are all acceptable, which is one of the main reasons for the gradual application of deep learning in spectral analysis.
| Datasets | Model | RAW | Stander | MSC | SG | SNV |
|---|---|---|---|---|---|---|
| Cefixime | SVM | 26.00 | 97.00 | 21.00 | 26.00 | 81.00 |
| PLS_DA | 20.00 | 95.00 | 20.00 | 20.00 | 67.00 | |
| SAE | 99.00 | 99.00 | 98.00 | 98.00 | 100.00 | |
| CNN | 98.00 | 100.00 | 100.00 | 100.00 | 99.00 | |
| SpectraTr | 99.00 | 100.00 | 100.00 | 100.00 | 99.00 | |
| Cefixime and phenytoin sodium | SVM | 21.59 | 85.39 | 18.73 | 21.59 | 50.04 |
| PLS_DA | 23.87 | 91.36 | 10.15 | 11.16 | 37.16 | |
| SAE | 86.03 | 99.28 | 76.82 | 85.71 | 98.41 | |
| CNN | 92.61 | 99.64 | 91.42 | 99.37 | 95.62 | |
| SpectraTr | 96.42 | 99.68 | 96.30 | 99.15 | 94.06 |
In this experiment, we observed that the SpectraTr model is less dependent on pre-processing than the CNN model. On cefixime, the accuracy of the original spectra-based SpectraTr model was 99%, with a difference of less than 1% from the prediction of the qualitative analysis model based on the pre-processing spectra. The raw spectral-based SpectraTr model achieves 96.42%, while the Stander, MSC, SG and SNV pretreatment models achieve 99.68%, 96.30%, 99.15%, and 94.06% on cefixime and phenytoin sodium, respectively. This demonstrates that the SpectraTr model can extract features from spectral data with various noise components automatically.
3.2. Result of dataset B
To compare with previous research effectively, we used the same open-source dataset as in Ref. 10 and conducted experiments with the arrangement described in the text. 201 samples were randomly selected to create the train set, and the remaining 99 samples were utilized as the test set. The prediction performance of the SpectraTr model on the original spectrum is shown in Table 7.
| Dataset | Acc | Precision | Recall | F1-score | CNN10 |
|---|---|---|---|---|---|
| Table | 96.97 | 97.03 | 96.97 | 96.98 | 94% |
The SpectraTr model achieve 96.97%, 97.03%, 96.97%, and 96.98% of accuracy, specificity, sensitivity, and F1-score, respectively, better than the model built based on CNN. The prediction performance of SpectraTr model and other baseline models under different pre-processing are shown in Table 8.
| Model | RAW | Stander | MSC | SG | SNV |
|---|---|---|---|---|---|
| SVM | 68.69 | 78.57 | 40.20 | 68.86 | 40.20 |
| PLS_DA | 62.12 | 79.58 | 47.20 | 60.25 | 55.25 |
| SAE | 91.92 | 95.95 | 62.26 | 92.93 | 58.59 |
| CNN | 94.24 | 98.21 | 90.77 | 95.54 | 90.53 |
| SpectraTr | 96.97 | 99.21 | 89.36 | 96.09 | 88.85 |
SpectraTr reaches 99.21%, 89.36%, 96.09%, and 88.85%, respectively after the pre-processing of standardization, MSC, SG, and SNV, which are better than other models in most cases. But pre-processing has a negative gain on SpectraTr in the vast majority of situations. We think that the deep learning method does not rely on pre-processing or only needs standardized pre-processing, because the deep learning method has good spatial invariance. The extensive application of the CNN algorithm in the image shows that image rotation, translation, and clipping have little effect on the prediction results of the deep learning model. In our previous studies, we have also found that the deep learning method has good adaptability to the slight rotation of spectra.31 Therefore, deep learning does not rely too much on preprocessing algorithms to remove noise such as baseline drift in spectra but needs to be standardized to allow faster gradient convergence.
3.2.1. The effect of hyperparameters on model performance
The above experiments have shown the qualitative analysis performance of the SpectraTr model and the ability of automatic feature extraction under different preprocessing. But the research shows that the network of the deep learning model is large and sensitive to the setting of network parameters. Therefore, on our drug and open-source drug data sets, the depth of the spectral encoder, the number of multi-heads, the number of patches, and the dimension of Mlp_dim are used as objects to study the impact of the above parameter settings on the spectralTr. The results are shown in Fig. 6.
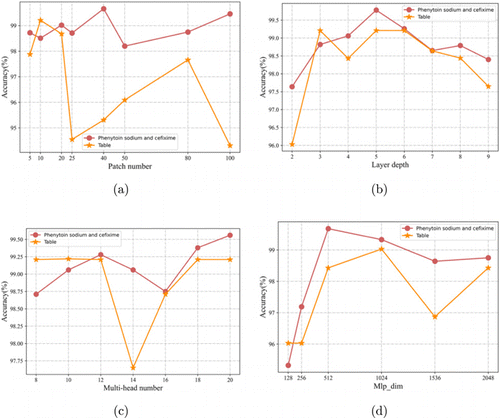
Fig. 6. The effect of model hyperparameters on perdition in dataset A and dataset B.
The effect of different patch numbers on the model is shown in Fig. 6(a). The difference in dataset A is not large. It is relatively better when the patch number is set to 40. As for dataset B, the performance is relatively best when the patch number is set to 10. When the patch number is set to 25, the prediction accuracy drops below 95%. However, due to the different spectral dimensions of dataset A and dataset B, all the results perform better when there are about 50 spectral points in the patch. Figure 6(b) shows the influence of Spectra encoder depth on the model. On the two data sets, the effects are similar. A network that is too shallow is not enough to extract features. If it is too deep, it is easy to overfit. From Fig. 6(c), the parameter setting of the number of attention heads has little effect on the model, and the model prediction accuracy fluctuates around 3% under different parameter settings. The influence of different parameters of Mlp_dim on the model performance is shown in Fig. 6(d), and the effect is better when it is around 512 or 1024. In summary, the four groups of main parameters can make the model have better predictive performance in a larger range, which shows that the SpectraTr model has low sensitivity to the parameter settings of the network layer. Therefore, when setting up the SpectraTr model, there is no need to search for the optimal parameter combination one by one from the massive parameter space. This has an advantage over the CNN model in modeling.
3.3. The result of training time
The training time of different models on datasets A and B is shown in Fig. 7. Different numbers of training samples will affect the model training time. The experiment of the table dataset was carried out when the number of training samples was 211, and the rest of the dataset was tested under the 8:2 training test set.
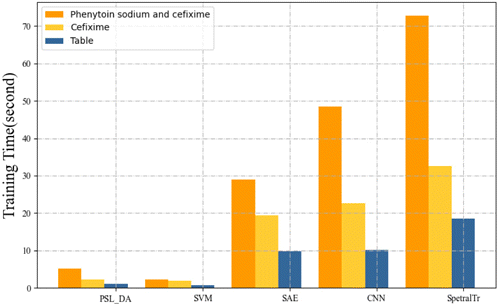
Fig. 7. The result of training time in datasets A and B.
The training period for deep learning is substantially longer than for SVM and PLS_DA algorithms since it takes several training sessions. The need for greater hardware resources and longer training time is the drawback of deep learning methods. The SpectraTr model has the longest training time among the deep learning models. The SpectraTr model’s training time is affected by both the number of samples and the spectral dimension. The training time increases as the number of samples and spectral dimensions increases.
4. Conclusion
A novel and advanced deep learning-based model is proposed called SpectraTr for qualitative analysis of near-infrared spectroscopy of drugs. Experimental results on multiple drug classification datasets show that SpectraTr outperforms the classical PLS, SVM, and existing deep learning methods AE and CNN in most cases. In addition, the model can automatically extract features from spectral data and does not rely on complex pre-processing and model parameter selection engineering. The initial application of SpectraTr in spectroscopy has shown potential and is expected to become a major class of deep learning methods in addition to AE networks and CNN networks.
Acknowledgments
The authors would like to express our gratitude to anonymous reviewers and editors for their helpful comments. This work was supported by the National Natural Science Foundation of China (61906050 and 21365008); Guangxi Technology R&D Program (2018AD11018), and Innovation Project of GUET Graduate Education (2021YCXS050).


