Construction of a multi-tissue compound–target interaction network of Qingfei Paidu decoction in COVID-19 treatment based on deep learning and transcriptomic analysis
Abstract
The Qingfei Paidu decoction (QFPDD) is a widely acclaimed therapeutic formula employed nationwide for the clinical management of coronavirus disease 2019 (COVID-19). QFPDD exerts a synergistic therapeutic effect, characterized by its multi-component, multi-target, and multi-pathway action. However, the intricate interactions among the ingredients and targets within QFPDD and their systematic effects in multiple tissues remain undetermined. To address this, we qualitatively characterized the chemical components of QFPDD. We integrated multi-tissue transcriptomic analysis with GraphDTA, a deep learning model, to screen for potential compound–target interactions of QFPDD in multiple tissues. We predicted 13 key active compounds, 127 potential targets and 27 pathways associated with QFPDD across six different tissues. Notably, oleanolic acid-AXL exhibited leading affinity in the heart, blood, and liver. Molecular docking and molecular dynamics simulation confirmed their strong binding affinity. The robust interaction between oleanolic acid and the AXL receptor suggests that AXL is a promising target for developing clinical intervention strategies. Through the construction of a multi-tissue compound–target interaction network, our study further elucidated the mechanisms through which QFPDD effectively combats COVID-19 in multiple tissues. Our work also establishes a framework for future investigations into the systemic effects of other Traditional Chinese Medicine (TCM) formulas in disease treatment.
1. Introduction
A novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused the global pandemic known as coronavirus disease 2019 (COVID-19).1 To date, SARS-CoV-2 has afflicted more than 700 million individuals worldwide and caused over 6 million fatalities.2 Clinical symptoms of COVID-19 patients include fever, fatigue, cough, shortness of breath, anorexia, and confusion.3,4 In severe cases, patients may die of respiratory failure, septic shock, or multiple organ failure.4 It is noteworthy that SARS-CoV-2 is capable of infecting and replicating within multiple organs throughout the body, including the lungs, heart, brain, and kidneys.5 SARS-CoV-2 enters human cells by binding to angiotensin-converting enzyme 2 (ACE2), which is also widely distributed across various organs, including the lungs, vasculature, heart, kidneys, and brain.6
Currently, antiviral drugs such as Nirmatrelvir/Ritonavir (PaxlovidTM), Remdesivir, and Molnupiravir hinder viral replication by inhibiting the main SARS-CoV-2 protease, Mpro or RNA-dependent RNA polymerase (RdRp).7 In contrast, traditional Chinese medicine (TCM) offers a multifaceted approach with multiple therapeutic targets for managing COVID-19. The use of TCM has effectively curbed the rapid spread of SARS-CoV-2 in China.8 One such TCM remedy is the Qingfei Paidu decoction (QFPDD), a blend of classical TCM formulations, including Maxing Shigan decoction, Shegan Mahuang decoction, Xiao Chaihu decoction, and Wuling powder. QFPDD has gained nationwide recognition and is endorsed by the National Health Commission and the State Administration of Traditional Chinese Medicine for the clinical treatment of COVID-19.9
QFPDD, a TCM formula consisting of 20 herbs and the mineral drug Gypsum Fibrosum (Sheng Shi Gao), is prepared as a decoction for oral administration. A meta-analysis has revealed that combination therapies incorporating QFPDD can significantly reduce nucleic acid conversion time and hospitalization duration.10 However, the widespread application of QFPDD beyond China has been hampered by its complex composition and unclear mechanism of action. Previous research has shown that QFPDD operates with a synergistic regulatory effect involving multiple components, targets, and pathways, making it challenging to pinpoint the most relevant targets due to the presence of numerous active ingredients.11,12,13 Moreover, the targets of QFPDD for the treatment of COVID-19 in different tissues remain uncharacterized.
In drug development, the foundation lies in understanding drug–target interactions (DTIs). Experimental screening for drug–target pairs is costly and time-consuming. Deep learning methods, including DeepPurpose14 and DeepDTA,15 provide enhanced capabilities for identifying, processing, and inferring complex patterns in molecular data, distinguishing them from traditional low-throughput techniques.16,17,18 Common deep learning-based DTI models consist of two essential components: the encoder and the decoder, providing a unifying mathematical framework for DTI prediction. Graph convolution algorithms, notably exemplified by GraphDTA,19 incorporate structural information of molecules and have demonstrated superior performance in computational drug discovery.20
In pursuit of a more comprehensive exploration of the active components of QFPDD and their relevant targets in multiple tissues for COVID-19 treatment, we qualitatively analyzed the chemical components of QFPDD using ultraperformance liquid chromatography plus Q-Exactive Orbitrap tandem mass spectrometry (UPLC-Q-Exactive Orbitrap-MS). We further integrated transcriptomic analysis with the GraphDTA model to enhance DTI prediction in multiple tissues. Our findings were subsequently validated through molecular docking, molecular dynamics simulations, and biological network analyses. This approach provided deeper insights into the mechanisms of QFPDD in treating COVID-19, leading to the establishment of a multi-tissue compound–target network and setting the stage for future experimental verification. The integration of data mining, deep learning techniques, and bioinformatic analysis equips us with the means to identify potential targets for disease treatment using TCM. A visual representation of our study process is depicted in Fig. 1.
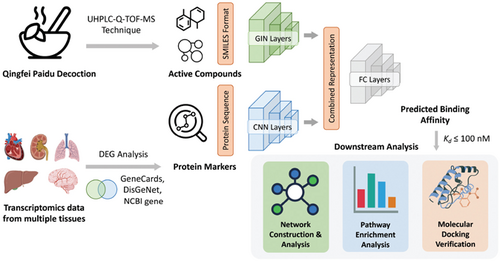
Fig. 1. The pipeline for constructing multi-tissue regulatory network of QFPDD against COVID-19. First, active compounds of QFPDD were determined using the UHPLC-Q-TOF-MS technique. Next, analysis of bulk transcriptomics revealed differentially expressed genes (DEGs) in each tissue type. We intersected DEGs in each tissue type with high-confidence COVID-19-related genes obtained from three public databases. The deep learning model GraphDTA was used to predict the binding affinity between active compounds and COVID-19-related proteins. Finally, compound–target interactions with high binding affinity were selected for downstream analysis, including network construction, pathway enrichment analysis, and molecular docking verification.
2. Materials and Methods
2.1. Materials and reagents for UPLC-Q-Exactive Orbitrap-MS analysis
The following materials and reagents were purchased for the qualitative analysis of the chemical components of QFPDD using UPLC-Q-Exactive Orbitrap-MS. The reference compounds, including amygdalin, ephedrine hydrochloride, saikosaponin A, quercetin, chlorogenic acid, glycyrrhizic acid ammonium salt, and 4,5-dicaffeoylquinic acid, were purchased from National Institutes for Food and Drug Control (Beijing, China). Hesperidin and irigenin were purchased from Shanghai Winherb Medical Technology Co., Ltd. (Shanghai, China). HPLC-grade acetonitrile, methanol, and formic acid were purchased from Merck (Darmstadt, Germany). Deionized water was prepared using a Millipore water purification system (Millipore, Merck, Darmstadt, Germany). All other reagents were of analytical purity.
QFPDD contained Herba Ephedrae (Ma Huang), Semen Armeniacae Amarum (Ku Xing Ren), Ramulus Cinnamomi (Gui Zhi), Rhizoma Alismatis (Ze Xie), Polyporus Umbellatus (Zhu Ling), Rhizoma Atractylodis Macrocephalae (Bai Zhu), Poria (Fu Ling), Radix Bupleuri (Chai Hu), Radix Scutellariae (Huang Qin), Rhizoma Pinelliae Praeparatum (Ban Xia), Rhizoma Zingiberis Recens (Sheng Jiang), Radix Asteris (Zi Wan), Flos Farfarae (Kuan Dong Hua), Rhizoma Belamcandae (She Gan), Herba Asari (Xi Xin), Rhizoma Dioscoreae (Shan Yao), Fructus Aurantii Immaturus (Zhi Shi), Pericarpium Citri Reticulatae (Chen Pi), Herba Pogostemonis (Guang Huo Xiang), Radix Glycyrrhizae (Gan Cao), and a mineral drug Gypsum Fibrosum (Sheng Shi Gao). These ingredients were purchased from China Beijing Tongrentang (Group) Co., Ltd.
2.2. QFPDD sample preparation
To prepare the QFPDD sample for UPLC-Q-Exactive Orbitrap-MS analysis,Gypsum Fibrosum (Sheng Shi Gao) was decocted for 15min initially. Then, each TCM ingredient was separately decocted twice with eight times and six times the amount of water, each time for 30min. The decoction liquids were combined, and filtered, and the sample solution was obtained. The sample solution was vortexed and mixed uniformly. 200μL of each sample was taken and crushed in a tissue disruptor with three steel beads. Then, 1000μL of pre-cooled methanol/water (8:2, v/v) was added, mixed, sonicated in an ice bath for 30min, allowed to stand at −20∘C for 60min, and centrifuged at 16,000g for 20min at 4°C. The supernatant was collected. The supernatant was evaporated to dryness in a high-speed vacuum concentrator, re-dissolved in 100μL of methanol-water solution (8:2, v/v), and centrifuged at 20,000g for 20min at 4°C. The supernatant was collected for injection analysis.
2.3. UPLC-Q-Exactive Orbitrap-MS analysis
The samples were analyzed using a Shimadzu LC-30AD UHPLC system (Shimadzu, Kyoto, Japan). Samples were separated on a Waters ACQUITY UPLC HSS T3 (2.1mm×100mm, 1.8μm) at a flow rate of 0.2mL/min and a column temperature of 40°, with an injection volume of 5μL. The mobile phases were as follows: A. 0.1% formic acid-water, and B. acetonitrile. The gradient elution procedure was as follows: 0–8min, 0–0% B; 8–45min, 0–40% B; 45–50min, 40–100% B; 50–60min, 100–100% B; 60–60.1min, 100–0% B; 60.1–70min, 0–0% B.
A QE Plus mass spectrometer (Thermo Fisher Scientific, CA, United States) with electrospray ionization (ESI) was used for mass spectrometry detection in both positive and negative ion modes. For Q-exactive Orbitrap-MS analysis, the source settings were as follows: spray voltage: 3.8kV (positive mode) and 3.2kV (negative mode), capillary temperature: 320°C, sheath gas: 30arb, auxiliary gas: 5arb, probe heater temperature: 350°C, S-Lens RF level: 50. Samples were analyzed in two scan modes: Full MS with a resolution of 70,000 and dd-MS2 with a resolution of 17,500, covering a scan range of m/z 70–1050. The system was regulated by Xcalibur software, and collected data were processed using MS-DIAL software version 4.21
2.4. Disease-related gene identification in multiple tissue types
We searched for COVID-19-related transcriptomic data of multiple tissue types from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Our criteria for selecting datasets were as follows: (1) explicit statement of COVID-19 cases and healthy controls without additional conditions; (2) gene expression profiling of human tissues; (3) in case where multiple datasets met criteria (1) and (2) for a certain tissue type, we selected the one with the largest sample size. See Table 1 for detailed descriptions of the datasets used.
| Study | Tissue type | n (case) | n (control) |
|---|---|---|---|
| GSE217948 | Whole blood | 203 | 71 |
| GSE205099 | Lung | 48 | 16 |
| GSE188847 | Brain | 22 | 23 |
| GSE202182 | Kidney | 13 | 10 |
| GSE169241 | Heart | 3 | 5 |
| GSE112356/GSE150316 | Liver | 3 | 3 |
We used the DESeq2 R package22 to screen for differentially expressed genes (DEGs) in each tissue type, using |logfoldchange|>=2 and adjusted P<0.05 as thresholds. To control false positive discoveries from DEG analysis, we intersected DEGs in each tissue type with high-confidence COVID-19-related genes obtained from three public databases: (1) DisGeNET (http://www.disgenet.org/),23 (2) GeneCards (https://www.genecards.org/),24 and (3) NCBI Gene (https://www.ncbi.nlm.nih.gov/).25 The intersected genes in each tissue type were visualized using VENNY2.1 (https://bioinfogp.cnb.csic.es/tools/venny/). The amino acid sequences corresponding to the identified protein-coding genes were then retrieved from the UniProt database.26
2.5. Drug–target interaction prediction methods
Graph isomorphic network (GIN) is recognized as one of the most robust neighborhood aggregation-based Graph neural networks (GNNs). This model takes the adjacency matrix, node feature matrix, and graph labels as inputs, generating embedded features of the graph through a readout layer. Notably, GIN excels in representing graphs effectively and demonstrates remarkable discriminative capabilities, making it particularly suitable for molecular classification tasks.27 GIN employs a multi-layer perceptron (MLP) to update node characteristics, and the GIN-based GNN consists of five GIN layers, each followed by a batch normalization layer :
GraphDTA is constructed by combining five GIN layers and three 1D convolutional neural network (CNN) layers, facilitating drug–target binding affinity prediction (https://github.com/thinng/GraphDTA).19 The model takes a vector representation of a protein and drug pair as input, yielding a continuous value that characterizes the affinity of the pair.
The drug–target binding affinity prediction model encompasses two inputs: SMILES strings and amino acid sequences. These inputs are fed into the GIN and CNN models, enabling them to learn the corresponding representation vectors. Subsequently, the two potential feature vectors are connected, and the affinities in the regression layers are estimated by two dense layers. SMILES strings are generated by transforming the 2D structures of molecules into linear structures based on established rules and then converting them into corresponding ASCII strings.28
For the GIN layers with the drug’s SMILES as input, the open-source software RDKit (https://rdkit.org/) was used to construct the molecular graph of the drug and extract the atomic features. Each drug SMILES was treated as a graph, and the resulting graph structure data were input into the GIN layer, allowing it to learn underlying patterns in the drug graph feature representation. This process led to the construction of a corresponding molecular graph that reflects the internal atoms of the interacting compounds. For the amino acid sequence input, the model treated the sequence as a string of ASCII characters. Three 1D CNN layers were applied to learn the sequence representation vector.
2.6. Training set, validation set and model hyperparameters
We used the 2020m2 version of the BindingDB dataset29 (https://www.bindingdb.org/bind/downloads/BindingDB_All_2020m2.tsv.zip), which includes four affinity scores: kinase inhibition constant (Ki), kinase dissociation constant (Kd), median effective concentration (EC50), and median inhibitory concentration (IC50), all measured in in nM. The Kd value was used for the model prediction task. The BindingDB dataset contains interactions between 12,184 targets and 1502 drugs, resulting in 69,479 pairs of affinity interaction results (measured by Kd value). The Davis kinase database30 contains selected assays of kinases and related inhibitors, along with their respective Kd values. The Davis dataset consists of interactions between 442 targets and 68 drugs, resulting in 30,056 pairs of affinity interaction results (measured by Kd value). Considering the sample sizes, we used the BindingDB dataset (measured by Kd value) as the training set and the Davis dataset (measured by Kd value) as the validation set.
Considering that the dissociation constant is non-normally distributed, we converted Kd to obtain pKd (pKd=−log). We used nM as a threshold to define the existence of DTI, following the convention generally used in the literature.31
We set the hyperparameters of the deep learning model as follows: For the GIN model, the feature number is 78, the output dimension size is 128, and the dropout is 0.2. For the CNN model, the feature number is 25, the filter number is 32, and the embedding size is 128. In the prediction model, the learning rate is 0.0005, the epoch number is 500, the training set batch size is 1024, the validation set batch size is 1024, and the test set batch size is 1024.
2.7. Model evaluation metrics
As the affinity scores are regression values, we used mean squared error (MSE), Pearson correlation coefficient, and concordance index (CI) as model evaluation metrics. A smaller MSE value indicates better accuracy in fitting the experimental data by the prediction model. The Pearson correlation coefficient reflects the degree of linear correlation between two random variables. CI assesses the prediction ability of the model by estimating the probability that the predicted results are consistent with the actual observed results.
2.8. Pathway enrichment analysis
Pathway enrichment analysis and visualization were performed using the ReactomePA,32 clusterProfiler,33 and enrichplot34 packages. For pathway enrichment analysis, the P values were calculated using a hypergeometric test. We used the gene concept networks to visualize the complex relationships between genes and the biological concepts they are enriched. All statistical tests were two-sided, and significance was considered at . All data analysis and visualization were performed using R version 3.5.0 (http://www.r-project.org/) and Python version 3.7.3 (https://www.python.org).
2.9. Network construction
To better visualize the complex interactions between potential active compounds and disease-related targets among multiple tissues, we constructed two types of networks using Cytoscape software (version 3.7.2)35: (1) protein–protein interaction (PPI) networks of target genes in various tissues; and (2) a tissue–compound–target (T–C–T) network demonstrating the interactions between potential active compounds and their target genes among six tissues.
2.10. Molecular docking
To further validate the binding between oleanolic acid and AXL and elucidate the underlying molecular mechanisms of their interaction, we performed molecular docking to model the binding interactions. The initial structure of AXL protein was obtained from the Protein Data Bank (PDB) database (https://www.rcsb.org/)36 with an accession code of 5u6b. The docking data were preprocessed using PyMOL software (open-source version), which involved extracting original ligands and deleting water molecules. Subsequently, we used AutoDock Tools (version 1.5.7) to prepare coordinate files in PDBQT format by adding polar hydrogen atoms and assigning partial charges. The 3D structures of these ligands were sourced from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and saved as files in SDF format. These files were then converted to pdb format using PyMOL software. The files in PDBQT format were prepared by adding polar hydrogen atoms, assigning partial charges, and defining rotatable bonds with AutoDock Tools (version 1.5.7). The substrate was docked into the active site of the protein using AutoDock Vina tool37 in Chimera.38 The binding site was set as a Å cube centered on the original ligand with a grid space of [24.4, −13.1, 17.1]. The docked poses with the highest docking scores were selected for subsequent molecular dynamics simulations.
2.11. Molecular dynamics simulation
Molecular dynamics simulations were conducted using the GROMACS 2022.4 package39 to investigate the dynamic interactions between oleanolic acid and AXL. The general AMBER force field (GAFF)40 was applied to substrates, and the partial atomic charges were obtained from the RESP method by Multwfn.41 The parmchk utility from AMBERTools was used to generate the missing parameters for the ligands. Na and Cl-ions were added to the protein surface to neutralize the total charges of the systems. Finally, the resulting systems were solvated in a rectangular box of TIP3P42 waters, extending up to a minimum cutoff of 15Å from the protein boundary. The Amber ff14SB force field43 was employed for the protein in all molecular dynamics simulations. The initial structures were fully minimized using the conjugate gradient algorithm, followed by 50,000 steps of steepest descent energy optimization. The systems were then gently annealed from 10 to 300K under a canonical ensemble for 0.05ns with a weak restraint of 15kcal/mol/Å. A 1-ns density equilibration was performed under an isothermal-isobaric ensemble at the target temperature of 300K and the target pressure of 1.0atm using Langevin-thermostat44 and Berendsen barostat45 with a collision frequency of 0.002ns and a pressure-relaxation time of 0.001ns. Further equilibration of the systems was allowed for 500ps to achieve a well-settled temperature and pressure. After proper minimizations and equilibrations, a 200ns production molecular dynamics simulation was performed to characterize the dynamics of the docked enzyme-substrate complexes. The root-mean-square deviation (RMSD) of the ligand backbone atoms was calculated along the molecular dynamics trajectories to explore the conformational stability of the binding poses.
3. Results
3.1. Characterization of chemical ingredients of QFPDD
We conducted a qualitative analysis of the chemical components of QFPDD using ultraperformance liquid chromatography plus Q-Exactive Orbitrap tandem mass spectrometry (UPLC-Q-Exactive Orbitrap-MS). A total of 83 compounds were identified in the positive ion mode (Fig. 2(a)), and 88 compounds were identified in the negative ion mode (Fig. 2(b)) by searching the SymMap database (http://www.symmap.org/), HERB database (http://herb.ac.cn/), and TCMSP database (https://tcmsp-e.com/), as well as by comparing them with literature and reference standards. After removing 15 duplicated compounds, a total of 156 compounds were identified (Supplementary Table S1). Nine compounds were confidently identified by comparing their retention times and mass spectrometry information with reference standards (Supplementary Fig. S1).
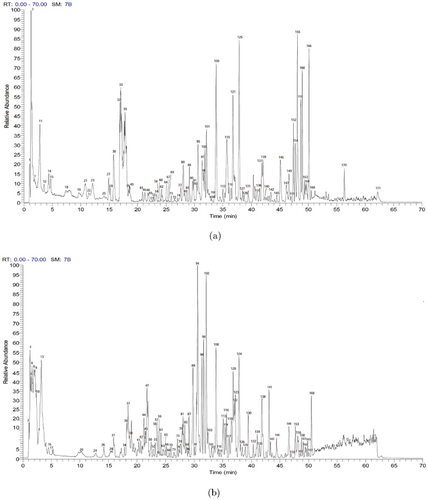
Fig. 2. Mass spectrum chromatograms of QFPDD. (a) Positive ion mode. (b) Negative ion mode.
3.2. Prediction of compound–target interactions
To identify potential protein targets of QFPDD in multiple tissue types, we performed differential gene expression analysis between COVID-19 patients and healthy controls in each tissue type independently. We identified 540 DEGs in the whole blood, 204 in the brain, 1358 in the heart, 1456 in the kidney, 2392 in the liver, and 1020 in the lung. Moreover, 3354 COVID-19-associated genes were obtained from 3 public databases: DisGeNET, GeneCards, and NCBI Gene. DEGs in each tissue type were then independently intersected with the 3354 COVID-19-associated genes, as visualized in Supplementary Fig. S2 using Venn diagrams. The intersected genes were matched to their corresponding proteins based on the Uniprot database.26 In total, we identified 127 potential target proteins in the whole blood, 83 in the brain, 266 in the heart, 308 in the kidney, 369 in the liver, and 200 in the lung. See Supplementary Table S2 for the full lists of COVID-19-associated genes and their corresponding proteins in each tissue type.
Predicting drug–target affinity (DTA) is beneficial for accelerating drug discovery, generating new therapeutic targets, and interpreting new mechanisms of drug therapy. Therefore, we predicted the affinity scores between 156 chemical ingredients of QFPDD and 127/83/266/308/369/200 COVID-19 candidate protein targets in six tissues: whole blood, brain, heart, kidney, liver, and lung. See Figs. 3(a) and 3(b) for the distribution of SMILES length and protein length. We used the GraphDTA model, employing publicly available datasets from BindingDB and Davis as the training set and validation set, respectively, for predicting the affinity scores. After fine-tuning the model and parameters, the validation set yielded the best prediction results with a MSE of 0.267, a Pearson correlation coefficient of 0.817, and a CI of 0.886 (Supplementary Table S3). The density plot showed that the maximum predicted pKd value was less than 10 and the peak pKd value was around 6. The pKd values vary slightly across different tissues but are within reasonable ranges (Fig. 3(c)). Based on the predicted affinity of nM, a total of 5988 (10.46%) high-confidence compound–target pairs were obtained, including 587 in the whole blood, 206 in the brain, 1450 in the heart, 1117 in the kidney, 1687 in the liver, and 941 in the lung. In total, 24 protein targets in the whole blood, 78 in the brain, 63 in the heart, 78 in the kidney, 76 in the liver, and 48 in the lung were involved in the high-confidence compound–target pairs (Supplementary Table S4).
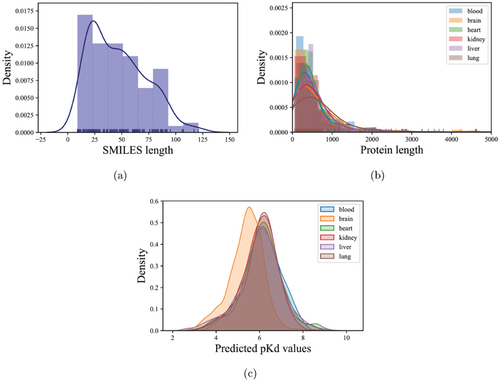
Fig. 3. Density distribution of SMILES length, protein length, and predicted pKd value in six tissues. (a) The canonical SMILES length of chemical ingredients of QFPDD. (b) Density plot of the protein length distribution in six tissues. (c) Density plot of the pKd results predicted by the GIN model in six tissues. Six tissues include brain, heart, kidney, liver, lung, and blood. Curves colored by blue, orange, green, red, pink, and brown represent the tissues of blood, brain, heart, kidney, liver, and lung, respectively.
3.3. Multi-tissue compound–target network of QFPDD in COVID-19 treatment
To identify hub protein targets, we constructed PPI networks for each tissue type by inputting tissue-specific protein targets into the STRING online database (https://string-db.org/cgi/input.pl)46 and visualizing the obtained data using Cytoscape35 (Supplementary Figs. S3(a)–S3(f)). To filter for hub protein targets, we applied a cutoff point based on the degrees within the PPI network. Consequently, hub protein targets were selected based on higher degrees and their central positioning within the PPI network. In each tissue type, we found the following number of hub protein targets: 10 proteins in whole blood, 36 proteins in the brain, 27 proteins in the heart, 43 proteins in the kidney, 28 proteins in the liver, and 17 proteins in the lung (Supplementary Table S5). A total of 127 protein targets were obtained excluding duplicates.
To identify potential key activate compounds that exert a broad impact across a large number of protein targets in various tissues, we first determined the total number of compound–target interactions for each compound across the six tissue types (Supplementary Table S6). We applied a cutoff point based on the summed number of interactions. This process allowed us to identify 13 key active compounds, including oleanolic acid, glycyrrhizin, formononetin glucoside, ursolic acid, 3’-Methoxypuerarin, 2-[4,5-Dihydroxy-6-[[8-hydroxy-8a-(hydroxymethyl)-4,4,6a,6b,11,11,14b-heptamethyl-1,2,3,4a,5,6,7,8,9,10,12,14a-dodecahydropicen-3-yl]oxy]-2-[[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxymethyl]oxan-3-yl]oxy-6-methyloxane-3,4,5-triol, niranthin, dehydroeburicoic acid, ephedrine, limonin, saikosaponin A, poricoic acid B, and uric acid. Together with the above-mentioned 127 hub protein targets, we constructed a tissue–compound–target (T–C–T) network, comprehensively demonstrating the multi-tissue compound–target interactions of QFPDD in COVID-19 treatment (Fig. 4).
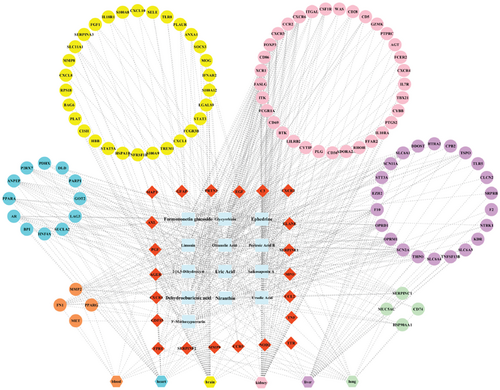
Fig. 4. The T–C–T network with 127 hub protein targets and 13 key active compounds. Hexagons represent tissues, ellipses represent targets, rectangles represent compounds, and diamonds represent targets common to different tissues.
3.4. Shared and unique pathways in QFPDD’s mechanism for COVID-19 treatment across tissues
Subsequently, we explored the pivotal pathways associated with the candidate targets of QFPDD in the context of COVID-19 treatment, using protein targets involved in high-confidence target-drug pairs from six tissues (Supplementary Table S4). Through Reactome pathway47 gene set enrichment analysis, we identified specific biological pathways associated with QFPDD treatment across these six tissues (Supplementary Table S7). Pathway enrichment analysis revealed 27 key pathways across these six tissues involved in the treatment of COVID-19 by QFPDD, including signaling by interleukins, neutrophil degranulation, chemokine signaling pathway, peptide ligand-binding receptors, and class A/1 (Rhodopsin-like) receptors (Fig. 5(a)). Specifically, several pathways were identified as shared key regulators in multiple tissue types, including interleukin-4 (IL-4) and interleukin-13 (IL-13) signaling in the blood, brain, kidney, liver and lung, as well as neutrophil degranulation in the blood, brain, heart, kidney, and lung (Fig. 5(a)). IL-4 and IL-13 are two cytokines that play an important role in regulating immune responses, especially in Th2-type immune responses. They activate a series of signal transduction pathways by binding to their respective receptors, regulating cell behavior and gene expression. In the context of COVID-19, the abnormal expression of these cytokines may impact the progression and severity of the disease.48 Thus, cytokine receptors, including the CCL, CCR, CXCR, FFAR, FPR, and GPR families, may play a crucial role in the treatment of COVID-19 by QFPDD in multiple tissues.

Fig. 5. Pathway enrichment analysis of the candidate targets for multiple tissue clusters. (a) The top 27 signaling pathways with high significance. The size of the dots represents the value of “GeneRatio” in each pathway, which denotes the number of genes annotated to a pathway within the specific list of differential genes among the major contributors that are included in the database. The color scale of the dots is based on the “P-value”. The “Cluster” denotes the number of all input genes for enrichment analysis in six tissues. Hypergeometric test was used in gene set pathway analysis. All statistical tests were 2-sided, and was considered significant. (b) The gene concept network involving multiple tissues and multiple pathways. The size of the circle indicates the number of genes in the pathway. Different tissues have different colors. (c)–(h) The top five significant biological pathways of each tissue: whole blood (c), brain (d), heart (e), kidney (f), liver (g), and lung (h). The red dots correspond to the pathway name, and the blue dots correspond to the gene name on the pathway. The size of the red dot indicates the number of genes enriched on the pathway.
By establishing a gene concept network, we showed that most of the top enriched pathways are interconnected across multiple tissues, serving diverse functions as receptors for various ligands, and including well-known drug targets (Fig. 5(b)). This provides compelling evidence for further experimental validation of candidate targets.
Considering the presence of unique biological pathway networks spanning multiple tissues, we conducted an analysis to identify the top five key pathways in each tissue. This analysis revealed that specific genes, such as BPI, GPR84, MPO, OLR1, PRTN3, ELANE, and MMP9 are primarily associated with neutrophil degranulation in the whole blood sample (Fig. 5(c)). Previous research has demonstrated that hypoxia-driven neutrophil degranulation may offer benefits for infection and inflammation.49 In the brain, major biological pathways include neutrophil degranulation and interleukin signaling, both of which affect numerous candidate targets (Fig. 5(d)). Interleukins exhibit pleiotropic effects on cells that bind them, influencing processes such as tissue growth and repair, hematopoietic homeostasis, and various levels of host defence against pathogens, making them an essential component of the immune system.50 In the heart, key biological pathways encompass neutrophil degranulation, glyoxylate metabolism and glycine degradation, nuclear receptor transcription pathway, pyruvate metabolism and citric Acid (TCA) cycle, and activation of AMPK downstream of NMDARs (Fig. 5(e)). The kidney exhibits highly active and intriguing biological processes, with Class A/1 (Rhodopsin-like) receptors and GPCR ligand binding standing out prominently (Fig. 5(f)). Class A/1 (Rhodopsin-like) receptors constitute the largest group of GPCRs, including chemokine receptors, and serving as the largest available group of drug targets. These receptors, including CCR2, CCR5, CXCR3, CXCR4, and CXCR6, bind to inflammatory chemokines that can direct leukocyte recruitment into infected tissues.51 Meanwhile, in the liver, enriched biological pathways primarily involve gamma-carboxylation of protein precursors, removal of aminoterminal propeptides from gamma-carboxylated proteins, gamma-carboxylation, transport, and amino-terminal cleavage of proteins, and regnenolone biosynthesis (Fig. 5(g)). Within the lung, important biological pathways include neutrophil degranulation, class A/1 (Rhodopsin-like) receptors, G alpha (i) signaling events, and formation of fibrin clot (clotting cascade) (Fig. 5(h)). Analysis of lung tissue sections revealed widespread extra- and intravascular compact fibrin deposits in patients with COVID-19, supporting the involvement of fibrin formation-related pathways.52
3.5. Verification of oleanolic acid-AXL binding through molecular docking and molecular dynamics simulation
Interestingly, we found that the interaction between oleanolic acid and the tyrosine-protein kinase receptor UFO (AXL) achieved the leading predicted affinity in multiple tissue types, including the heart, blood, and liver. Previous research has demonstrated that AXL is a candidate receptor for SARS-CoV-2 to enter human cells.53 Thus, the strong binding affinity between oleanolic acid and AXL may implicate potential clinical intervention strategies and wait for further experimental verifications. Here, we utilized molecular docking and molecular dynamics simulation to provide further computational support for the oleanolic acid-AXL binding.
Molecular docking indicated robust complex formation between oleanolic acid and AXL, characterized by a Gibbs free energy of −8.9kcal/mol. This high-affinity association can be primarily attributed to favorable electrostatic interactions between the ligand and key protein residues, including Leu542, Val550, Phe622, and Met679 within the binding pocket (Figs. 6(a) and 6(b)). Additionally, short-range van der Waals forces mediated by residues such as Gly545 and Asp627 contribute to stabilizing effects (Fig. 6(a)). In summary, the ligand-protein complex is stabilized by a combination of long-range electrostatic attractions to specific pocket residues, as well as short-range noncovalent interactions with nearby amino acids.
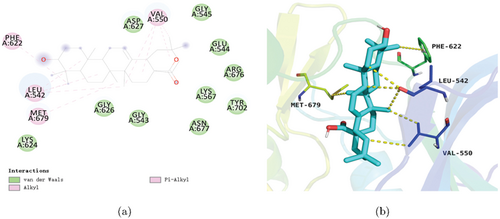
Fig. 6. Molecular docking pattern of oleanolic acid with the target AXL. (a) 2D action mode of oleanolic acid with the target AXL. (b) 3D action mode of oleanolic acid with the target AXL.
Moreover, structural characterization revealed that the apo-protein exhibits a volume of 54.36nm3 and a density of 983.18g/L. Upon ligand binding, the volume expands to 57.87nm3, while the density decreases to 964.46g/L in the complex. This subtle increase in volume and the concomitant reduction in density are consistent with a slight swelling of the protein after accommodating the ligand within the binding pocket. The volumetric and density changes support a model in which ligand engagement leads to localized alterations in protein shape and packing, enabling a high-affinity association.
Molecular dynamics simulations revealed a total of 225 hydrogen bonds between protein main and side chains in the unbound state (Fig. 7(a)), decreasing to 218 in the protein-ligand complex (Fig. 7(b)). Over the 200ns trajectories, the ligand formed a stable 2–3 hydrogen bond network with the protein (Fig. 7(c)). This reduction in overall hydrogen bonding upon ligand binding implied that the protein became partially destabilized and unfolded, likely due to strong interactions with the bound ligand that disrupt native contacts. These computational data suggested that the ligand acts as an inhibitor, perturbing the folded conformational ensemble and promoting partial protein denaturation.
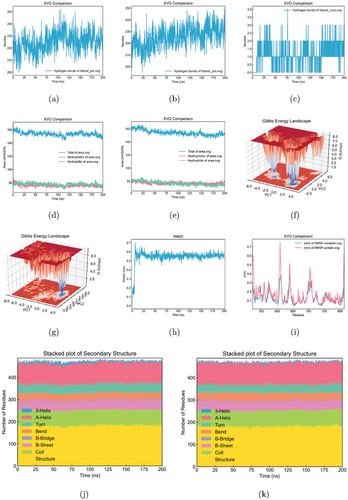
Fig. 7. Molecular dynamics simulations of oleanolic acid-AXL interaction. (a) Hydrogen bonds between AXL protein main and side chains in the unbound state. (b) Hydrogen bonds between AXL protein main and side chains in the AXL-oleanolic acid complex. (c) Hydrogen bonds between AXL and oleanolic acid in the AXL-oleanolic acid complex. (d) Solvent accessible surface area (SASA) of the unbound AXL protein. (e) SASA of the AXL-oleanolic acid complex. (f) The Gibbs free energy landscape of AXL protein. (g) The Gibbs free energy landscape of the AXL-oleanolic acid complex. (h) The RMSD plot of the AXL-oleanolic acid complex. (i) The RMSF plot of AXL protein and AXL-oleanolic acid complex. (j) The secondary structure conformation of the unbound AXL protein. (k) The secondary structure conformation of the AXL-oleanolic acid complex.
The analysis of solvent accessible surface area (SASA) revealed that the unbound protein exhibited a value of 153.82nm2 (Fig. 7(d)), increasing to 155.29nm2 when forming a complex with the ligand (Fig. 7(e)). This expansion in SASA implies greater solvent exposure of previously buried core residues, indirectly indicating a high-affinity interaction between the ligand and the protein. Furthermore, computed free solvation energies of 284.25 (kJ/mol × nm2) for the apo-protein increase to 296.46 (kJ/mol × nm2) for the complex, reflecting favorable energetic contributions from solvation upon ligand binding. In summary, the enhanced surface area exposure and solvation energy of the protein-ligand complex pointed to a high-affinity association driven by favorable desolvation and engagement of previously occluded binding site residues.
The conformational energy profile depicted the lower energy native state of the protein as a dark blue basin. The thermodynamically unfavorable transition from this dark to the light blue region signified that the global minimum was occupied on the energy landscape, appearing as a sharp, single low-energy state (Fig. 7(f)). However, upon binding, the ligand remodeled the landscape, shifting this global minimum. Ligand interactions can alter the Gibbs free energy surface, thus populating alternative conformations and generating distinct landscapes dependent on the bound ligand’s structure and mode (Fig. 7(g)).
The RMSD plots indicated that the complexes reached equilibrium after 5ns, with the ligand RMSDs converging around 6.0Å, suggesting stable binding orientations (Fig. 7(h)). Analyses of per-residue root-mean-square fluctuations (RMSF) revealed stabilized backbone dynamics for residues 600–620 of the protein when in the complex, likely due to binding interactions, consistent with the free energy landscapes (Fig. 7(i)). Notably, there were no significant changes in the protein’s secondary structure between the bound and unbound states, implying limited conformational changes upon binding for these systems (Figs. 7(j) and 7(k)).
Taken together, molecular docking and dynamic simulations provided valuable insights into the high-affinity binding of the oleanolic acid to the AXL protein, revealing structural and energetic changes that support a model of localized protein shape alterations and partial unfolding upon ligand engagement.
4. Discussion
According to TCM theory, COVID-19 is a damp-heat lung plague caused by damp-heat and epidemic toxins. QFPDD can relieve cough, clear heat, eliminate dampness, and eliminate toxins in the treatment of COVID-19. Based on the clinical observation of COVID-19 cases, QFPDD could effectively relieve symptoms such as fever, cough, asthma, expectoration, and nasal obstruction. The effective rate is higher than 80% with different dosage forms and different courses of treatment.54 Previous studies have demonstrated that QFPDD exerts a synergistic regulatory effect, characterized by its multi-component, multi-target, and multi-pathway action,55,56 which is supported by the conclusions of our study. We further investigated the intricate interactions among the herbal ingredients of QFPDD and their potential targets in multiple tissues, which remain undetermined in previous studies.
SARS-CoV-2 infection is a pathological process accompanied by excessive inflammation and immunodeficiency.57 SARS-CoV-2 enters host cells by binding to the ACE2 receptor, which is widely distributed in human tissues, including the lung, vasculature, heart, kidney, and brain,1 causing multiple organ damage. QFPDD exhibited high affinity for ACE2 in treating COVID-19 and exerted therapeutic effects by acting on multiple organs and tissues.58 Moreover, studies have shown that after oral administration, the active compounds of QFPDD were rapidly distributed in various tissues, including the blood, heart, liver, lung, kidney, and brain.59 However, previous studies on the efficacy and mechanism of action of QFPDD only focused on a single organ or tissue, such as the blood60 and the lung.61,62 Moreover, in previous studies, putative targets of TCM compounds were obtained from public databases such as DrugBank, STITCH, ETCM,11 and SwissTargetPrediction.13 However, these databases cannot distinguish disease-related proteins by tissue types. Instead, we obtained the samples from multiple tissues of COVID-19 patients from GEO database, and then integrated multi-tissue transcriptomic analysis with the GraphDTA model to predict the targets. By constructing a QFPDD multi-tissue compound–target interaction network, we addressed the difficulty of comprehensively assessing the synergistic regulatory effects of QFPDD in the treatment of COVID-19 and elucidated the mechanisms through which QFPDD effectively combats COVID-19 in multiple tissues.
As a result, 156 compounds of QFPDD were identified by UPLC-Q-Exactive Orbitrap-MS. Thirteen key active compounds, 127 potential targets, and 27 pathways associated with QFPDD across 6 different tissues were predicted. Oleanolic acid-AXL exhibited leading affinity in the heart, blood, and liver. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells.53 AXL is present in many tissues of the body except neural tissues,63,64 which is consistent with our research. Oleanolic acid exhibited antiviral,65 anti-inflammatory,66 antibacterial,67 and cardioprotective activities.68 Our Study, based on deep learning, molecular docking, and molecular dynamics simulations, suggested that QFPPD, especially its oleanolic acid component, may prevent the infection and replication of SARS-CoV-2 by inhibiting AXL activation.
Previous studies have shown that QFPDD may have a therapeutic effect on COVID-19 by exerting an anti-inflammatory effect,13,69,70 which is consistent with our conclusion. But the specific pathways through which QFPDD exerts its therapeutic effects vary across different studies. QFPDD could exhibit immune regulation and anti-inflammation through the HIF-1 signaling pathway, the Toll-like receptor signaling pathway,13 the renin-angiotensin system signaling pathway,69 and the sphingolipid signaling pathway.70 In our study, the pathway enrichment analysis identified interleukin-4 and interleukin-13 signaling as key regulators in the blood, brain, kidney, liver and lung in the treatment of COVID-19 by QFPDD. IL-4 and IL-13 are key cytokines that initiate potent type 2 inflammatory processes.71 IL-13 is a driver of pulmonary injury and COVID-19 severity.72,73 Neutrophil degranulation was the top enriched biological pathway shared in the blood, brain, heart, kidney, and lung of QFPDD. As important immune cells, neutrophils migrate to the site of inflammation, guided by chemokines, and then kill pathogens through respiratory bursts and degranulation.74 Our analysis suggested that QFPDD may exert its anti-inflammatory effects in multiple tissues through these shared pathways.
In addition, QFPDD has its own unique biological pathways in various tissues, which has not been investigated by previous studies. In the heart, key biological pathways encompass glyoxylate metabolism, pyruvate metabolism and citric acid (TCA) cycle, which are crucial processes for intracellular energy conversion to maintain the normal life activities and metabolic processes of cells. Among them, the TCA cycle in the mitochondria is the main way for the body to obtain energy. About one-third of hospitalized COVID-19 patients are accompanied by myocardial injury.75,76 Subunit S1 of the spike protein of SARS-CoV-2 might impair mitochondrial function in human cardiomyocytes by binding to ACE2.77 The combination of QFPDD with western medicine tended to mitigate the extent of heart impairment.78 In addition to its anti-SARS-CoV-2 and anti-inflammatory effects,79,80 QFPDD may also play a role in organ protection by repairing myocardial damage through biological pathways such as the TCA cycle. As the active ingredient in QFPDD, oleanolic acid may have played a role in its cardioprotective properties.68 In the liver, the primary enriched biological pathway involved the gamma-carboxylation of protein. The synthesis of coagulation factors in the liver requires various glutamate residue-containing proteins, which must first be carboxylated to gamma-carboxyglutamic acid (Gla)-containing proteins in order to be coagulatively active.81 COVID-19 is known to cause blood clotting disorders, and SARS-CoV-2 can enter the bloodstream via the respiratory route.82 Therefore, coagulation factor-related pathway in the liver may play a role in COVID-19 infection. As the active ingredient in QFPDD, oleanolic acid has been shown to demonstrate anticoagulant activities by inhibiting the expression of tissue factors.83
There are some limitations to our study. The drug–target binding affinity prediction model played an essential role in establishing the compound–target interaction network in our study. We used SMILES to partially represent the structural information and atomic composition of molecules, but it may not capture all the topology information. Furthermore, SMILES strings are context-dependent, meaning that two molecules with similar SMILES strings may have distinct structures. It has been shown that generating molecules solely based on SMILES strings cannot guarantee 100% chemical validity unless intricate constraints are incorporated.84 Molecular graphs naturally and intuitively represent the topological structure of molecules.85 They depict atoms as nodes, bonds as edges, and incorporate atom types and bond types as node and edge features, respectively. We suggest the development of DTI prediction models based on molecular graphs instead of SMILES strings, which may benefit the construction of more accurate compound–target interaction networks in similar studies.
5. Conclusion
In summary, through the construction of a multi-tissue compound–target interaction network, our study identified the multi-tissue targets of action in the treatment of COVID-19 with QFPDD, and elucidated that the underlying mechanisms of QFPDD against SARS-CoV-2 mainly involve anti-inflammatory and immunomodulatory action, as well as organ protection. The integration of deep learning and bioinformatic analysis equips us with the tools to identify active ingredients and candidate targets for treating various diseases with TCM. Our work also establishes a framework for future investigations into the systemic effects of other TCM formulas in disease treatment. By taking into account synergies between multiple tissues, our findings reveal that oleanolic acid exhibits a notably strong predicted binding affinity with the tyrosine-protein kinase receptor UFO, also known as AXL, across various tissue types, including heart, blood, and liver. The robust interaction between oleanolic acid and the AXL receptor suggests that AXL is a promising target for the development of clinical intervention strategies.
Abbreviations
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019; QFPDD, Qingfei Paidu decoction; TCM, traditional Chinese medicine; DTI, drug–target interaction; SMILES, simplified molecular input line entry system; DEG, differentially expressed gene; DTA, drug–target affinity; MSE, mean squared error; CI, concordance index; PPI, protein–protein interaction; SASA, solvent accessible surface area; RMSD, root-mean-square deviation; RMSF, root-mean-square fluctuation; GEO, Gene Expression Omnibus; GIN, graph isomorphic network; CNN, convolutional neural network; MLP, multi-layer perceptron; , kinase inhibition constant; , kinase dissociation constant; EC50, median effective concentration; IC50, median inhibitory concentration; PDB, Protein Data Bank; ACE2, angiotension-converting enzyme 2; AXL, tyrosine-protein kinase receptor UFO; TCA, tricarboxylic acid; PDH, pyruvate dehydrogenase.
Acknowledgments
We thank all the participants in this study. Xia Li, Xuetong Zhao, and Xinjian Yu are contributed equally to this work. This work was supported by a grant from the medical scientific research project on COVID-19 of the Leading Science and Technology Program of the Health Commission of Shanxi Province (No. 2021XM47), and Evidence-based Medicine Innovation Team for Integrating Chinese and Western Medicine to Prevent and Treat Lung Disease of Shanxi University of Chinese Medicine (No. 2022TD1008).
Supplementary Materials
The Supplementary Materials are available at: https://www.worldscientific.com/doi/suppl/10.1142/S0219720024500161
ORCID
Xia Li  https://orcid.org/0000-0002-5432-1665
https://orcid.org/0000-0002-5432-1665
Xuetong Zhao  https://orcid.org/0000-0002-3019-8615
https://orcid.org/0000-0002-3019-8615
Xinjian Yu  https://orcid.org/0000-0003-3938-5924
https://orcid.org/0000-0003-3938-5924
Jianping Zhao  https://orcid.org/0000-0002-3880-1255
https://orcid.org/0000-0002-3880-1255
Xiangdong Fang  https://orcid.org/0000-0002-6628-8620
https://orcid.org/0000-0002-6628-8620
Xia Li is an Associate Professor at the Third Clinical College, Shanxi University of Chinese Medicine. She received her Doctoral degree from the Hubei University of Chinese Medicine. Her research interests include Chinese medicine and bioinformatics.
Xuetong Zhao received her Doctoral degree from the CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Science and China National Center for Bioinformation; University of Chinese Academy of Sciences. Presently, she works at the National Genomics Data Center in Beijing. Her research focuses on genomics data mining.
Xinjian Yu is a Ph.D. candidate at the Baylor College of Medicine. Her research interests include bioinformatics and precision medicine.
Jianping Zhao is a Professor at the Third Clinical College, Shanxi University of Chinese Medicine. His research interests include Chinese medicine and pharmacognosy.
Xiangdong Fang is a Professor at the CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Science and China National Center for Bioinformation; University of Chinese Academy of Sciences. His research interests include clinical omics and translational medicine for stem cells and complex diseases.