Deep-learning-based methods for super-resolution fluorescence microscopy
Abstract
The algorithm used for reconstruction or resolution enhancement is one of the factors affecting the quality of super-resolution images obtained by fluorescence microscopy. Deep-learning-based algorithms have achieved state-of-the-art performance in super-resolution fluorescence microscopy and are becoming increasingly attractive. We firstly introduce commonly-used deep learning models, and then review the latest applications in terms of the network architectures, the training data and the loss functions. Additionally, we discuss the challenges and limits when using deep learning to analyze the fluorescence microscopic data, and suggest ways to improve the reliability and robustness of deep learning applications.
1. Introduction
Recent developments of super-resolution fluorescence microscopy allow researchers to overcome the Abbe diffraction limit,1 and it has been considered a promising technique to bring “optical microscopy into the nanodimension”. The great improvement in spatial resolution immediately opens the door to a variety of discoveries at the nanoscale.2 As an integral member of super-resolution microscopy methods, stimulated emission depletion (STED) microscopy3 employs a second laser beam that has a doughnut-shaped focal intensity distribution with zero intensity at the center, and its overlap with the excitation laser beam results in a smaller residual fluorescence spot, thereby sharpening the point spread function (PSF) and improving resolution.4 Single-molecule localization microscopy (SMLM), including stochastic optical reconstruction microscopy (STORM)5 and fluorescence photo-activated localization microscopy (PALM),6 first labels specific proteins or oligonucleotides with a fluorophore or an autofluorescent protein to light up those molecules of interest,7 and then constructs super-resolution image by localizing and merging the positions of molecules.8 Structured illumination microscopy (SIM)9,10 surpasses the optical diffraction limit twofold by applying varying, nonuniform illumination on samples and using dedicated computational algorithms to derive super-resolution information from sequentially acquired images.11
However, intense illumination and multiple acquisitions are required to produce a single super-resolution image. Computation is thus becoming increasingly crucial for the imaging process,12 since it decides what information should pass through the optical system. For instance, computational algorithm is applied to improve the resolution of diffraction-limited confocal microscopy images to match the resolution acquired with STED microscope, reducing the dependence on relatively sophisticated optical setups.13 In the case of STORM, several algorithms have been developed to cater for localizing closely spaced molecules, which can improve temporal resolution by increasing the density of activated fluorophores in each raw image and requiring fewer raw images.14 Reference 15 uses efficient algorithm to obtain a five-fold reduction in the number of raw images required for super-resolution SIM, and generate images under low light and short exposure time conditions, thereby reducing photobleaching. While successful application of conventional algorithms has been demonstrated, most of them suffer from two fundamental drawbacks: undesirably long data-processing time and insufficient image quality. Hence, any substantial improvements in the algorithms used for resolution enhancement or reconstruction are desired to decrease photobleaching and increase resolution.
Deep learning, as a data-driven approach, has been applied successfully to image processing in a variety of research fields, including image classification,16 segmentation,17,18 computed tomography,19 optical microscopy20 and other areas.21,22,23,24 Deep learning is a class of machine learning technique, which has more complex ways of connecting layers and larger amount of computing power in training than previous networks. Another remarkable advantage that deep learning has over conventional machine learning algorithms is automatic feature extraction.25,26 More importantly, deep neural networks can learn end-to-end image transformations, which is different from classical optimization approaches that need explicit analytical modeling and prior knowledge. All these advances drive deep learning closer to its original goal of artificial intelligence (AI), bringing about significant breakthroughs in solving biomedical problems. Super-resolution fluorescence microscopy, too, has recently benefited from deep learning to improve resolution or to reconstruct high-resolution images. Despite the limited amount of published research, these studies demonstrate infinite possibilities of deep learning in super-resolution microscopy.
This review paper aims to give an overview of the applications of deep learning in super-resolution fluorescence microscopy. In Sec. 2, the deep learning architectures proposed in previous publications for processing fluorescence microscopic images are presented. Section 3 compares and analyzes the application of these models. Section 4 discusses the current challenges and potential methods for overcoming these challenges. Lastly, an overall summary is given in Sec. 5.
2. Deep Learning Models
This section introduces convolutional neural network (CNN) and generative adversarial network (GAN) that are commonly employed to analyze fluorescence microscopic data, due to their superb performance in image reconstruction and resolution enhancement.
2.1. Convolutional neural network
A convolution is defined as a mathematical operation describing how to merge two sets of information.25 As its name implies, CNN generates a feature map by convolving the input data with trainable kernels or weights.27 Convolution layers and pooling layers are two typical processing layers in CNN.
At one position, the convolution layer first computes the matrix dot product between the input image and the convolution kernel, and then sums all the values to obtain the value at the corresponding entry of the output feature map. The convolution kernel is applied in a sliding window manner according to the stride size, and the orientation is generally from top to bottom and from left to right. The convolution operation is considered complete when the convolution kernel is slid across the entire input data. The convolution layer is followed by an activation function, such as the rectified linear units (ReLU),28 which is important in CNN and introduces a nonlinearity to the model. The properties of the feature map are related to the user-defined hyperparameters, like convolution kernel size, depth, stride size and zero padding.
Pooling layers are inserted between successive convolution layers to reduce the spatial dimension of the feature representation and to prevent overfitting.25 Max pooling and average pooling perform down-sampling operation by taking respectively the maximal element and the average value in the corresponding region and discarding redundant information. This is beneficial for the neural network,29 since pooling layers reduce the parameter numbers and improve computational efficiency. Both kinds of pooling can help to make the representation approximately invariant to small translations of the input.30 This invariance to local translation is useful, especially if we are more interested in whether some features are present than where they are.30
Unlike fully connected neural network, CNN utilizes two important properties including local connectivity and parameter sharing. The former is inspired by the biological visual system. Neurons in a convolution layer are only connected to a local region of the previous layers along the spatial dimensions, and the hyperparameter called the receptive field that controls how much of the width and height to be affected. Local connectivity allows us to extract features while reducing the parameter count per layer we need to train.25 Parameter sharing is another scheme to control the total number of parameters by sharing weights for all neurons in a two-dimensional slice of depth. Figure 1 compares full connections and local connections with parameter sharing. Every connection between the input unit () and output unit () in Fig. 1(a) has its own weight, specified by different weight , and there is no parameter sharing. In contrast, Fig. 1(b) uses the same three weights (with different colors for distinction of ) repeatedly across the entire input. It is clear that local connectivity with parameter sharing can significantly decrease the complexity of the model, and the number of weights is reduced from 25 to 3 in Fig. 1, thereby improving the computational efficiency of the network.
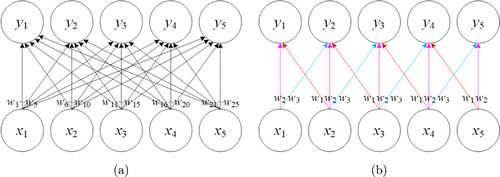
Fig. 1. Comparison of (a) full connections and (b) local connections. Each output unit is obtained by connecting different input units with corresponding weight . Arrows labeled with indicate the connections that are associated with a particular weight parameter. (a) Full connections have no parameter sharing so each weight is used only once. (b) Local connections use the weights repeatedly at all input locations due to parameter sharing.
In general, back-propagation is performed to train CNN,29and the gradient-based optimization algorithms, such as stochastic gradient descent (SGD)31 and the adaptive moment estimation (Adam) optimizer32 are employed to update the weights by minimizing the loss function. CNNs have been developed in fluorescence microscopy for super-resolution image reconstruction. These applications are further discussed in Sec. 3.
2.2. Generative adversarial network
Generative adversarial network (GAN) has attracted extensive attention since it was proposed by Ian Goodfellow in 2014.33 GAN can be divided into two parts, a generator and a discriminator. Figure 2 gives an illustration of the generative adversarial network and its training process, which is applied to enhance the image resolution. The generator is responsible for generating images from the input data while the discriminator estimates the probability of the generated output image being real. The goal here is to generate more reliable output images based on the training data, so that the discriminator cannot distinguish the images from the training set or from the generator.
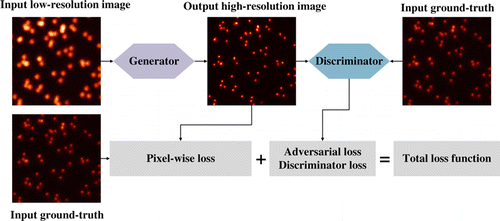
Fig. 2. Illustration of a generative adversarial network and its training process. The generator enhances the input low-resolution image, and the discriminator returns an adversarial loss to the output high-resolution image. The loss function can be designed as the combination of the adversarial loss with regularization terms.
GAN enables the generator and discriminator to compete with each other till they reach an equilibrium point. During the training, GAN works by alternating between training the generative model given the discriminative model, and updating the discriminator by keeping the generator unchanged. This adversarial training is achieved by optimizing the loss function with back-propagation technique. GAN training suffers from model collapse and gradient vanishing. One common failure mode involves the generator collapsing to produce a single sample or only a few similar samples of the true distribution.34 Additionally, the more quickly the discriminator learns to distinguish between real and fake images, the less reliable gradient information is provided to the generator. This results in vanishing gradient.35 Several algorithms have been proposed to address the above issues and to improve the stability of learning.36,37,38 Generative adversarial networks are potentially used for fluorescence microscopy due to their capability of enhancing image resolution, and their applications are elaborated in Sec. 3.
3. Deep Learning for Super-Resolution Fluorescence Microscopy
In the past few years, super-resolution fluorescence microscopy has experienced a rapid development. In particular, deep learning algorithms have shown promising results ranging from image reconstruction to image resolution enhancement. Deep learning is able to learn from data, and the design of proper network architecture, training data and loss function are crucial for its applications. Accordingly, this section introduces these three aspects of applications of deep learning to super-resolution images.
3.1. Stimulated emission depletion microscopy
The underlying principle of STED is selective deactivation of fluorophores by stimulated emission depletion to sharpen the point spread function. Deep learning methods can be applied to achieve image transformation between different imaging modalities and resolution enhancement.
Wang et al. present a deep-learning-based framework to achieve super-resolution and cross-modality image transformations in fluorescence microscopy.13 They transform diffraction-limited confocal microscopy images to match the resolution acquired with STED microscope. Although this work does not provide demonstration of its capability to directly super-resolving live-cell confocal images, it shows potential that the deep learning method is able to infer super-resolved live-cell images by training the model with only the static images. This can effectively overcome the constraints of photobleaching and phototoxicity on the practical long-term live-cell imaging with STED.
The network architecture used in this work is a conditional GAN model that has been proven to be quite effective in learning such transformation, where the input and output distributions share a high degree of mutual information.39 The generator is a CNN structure similar to the U-net, and the discriminator is also a simple CNN architecture. U-net has been applied successfully to suppress irrelevant regions while highlighting salient structures of varying shapes, yielding improved prediction performance across diverse datasets.40
The loss function of the generator is designed as the combination of the adversarial loss with the mean square error (MSE) and the structural similarity (SSIM), simultaneously ensuring prediction accuracy and perceptual quality fidelity. The loss function of the discriminator calculates the binary cross-entropy (BCE). They are formulated as
For the training data, since corresponding confocal image and STED image pairs are used to train the model, accurate registration and alignment are necessary and crucial as a pre-process step. This can reduce possible artifacts. It is also noticed that the network model should be retrained for optimal results when applying the approach to new types of samples unseen in the training stages. Therefore, the authors employ the transfer learning,41 which uses a previously trained network for another type of sample as the initial model, and speeds up the convergence of the learning process for new sample. The final model is selected with the smallest validation loss, which takes 90h to train. After transfer learning, it takes 24h to train the model with 8 patches ( pixels).
3.2. Single-molecule localization microscopy
The basic principle of SMLM is the localization of sparsely distributed fluorescent proteins/fluorophores over thousands of frames. Computational image reconstruction algorithms are required to detect precisely the position of the individual molecule.
3.2.1. Stochastic optical reconstruction microscopy
Nehme et al. present a deep convolutional neural network used for localization microscopy (Deep-STORM).42 It not only achieves state-of-the-art resolution under different signal-to-noise ratio (SNR) conditions and high molecule densities, but also shows significantly higher speed than existing approaches. Because STORM generally requires thousands of frames of raw images to reconstruct a super-resolution image, long acquisition time and time-consuming reconstruction algorithm have become a bottleneck that limits the wide adoption of STORM for live-cell imaging.14 As a consequence, fast and accurate reconstruction algorithms are desired for STORM. Deep-STORM pioneers in applying deep learning to reconstruct STORM data, demonstrating its feasibility and reliability.
The network architecture is based on a fully convolutional encoder–decoder network. The input image is firstly encoded into a dense and aggregated feature representation, and then reconstructed to a super-resolved image in the decoding stage. Both parts are constructed using CNN. Batch normalization (BN) is inserted between the convolutional layer and ReLU activation function to alleviate the internal covariate shift and to facilitate the training process.43
The loss function consists of a data-fidelity term and one regularization term. For the former, this work measures the squared norm of the difference between the network’s prediction and the desired ground-truth image convolved with a small 2D Gaussian kernel . Here, the regularization term is taken to control sparsity by penalizing the norm of the network’s output . Assuming that the number of images in the training set is N, the loss function can be described as
The training data are pairs of diffraction-limited images and super-resolved images with spikes at corresponding ground truth positions. Deep-STORM can be trained on simulated data or experimental data, and the simulated training examples can be generated by ImagJ44 ThunderSTORM45 plugin. To achieve the best reconstruction results, it is important to note that the parameters of the simulated training data should in principle be matched to the experimental conditions as close as possible, including the pixel size, the range of full width at half maximum (FWHM) of the point spread function (PSF), intensity range of the molecules and others. One prominent advantage of using simulated training data is that the amount and the quality of the data and their suitability are sufficient. The problem of using simulated training data will be further explored in Sec. 4. Full network training takes 2h with 7K pairs of training dataset ( pixels).
In another application of deep learning to reconstruct STORM data, Yao et al. focus on developing a reconstruction algorithm suited for high-density molecule localization and live-cell STORM imaging.8 This work designs a two-part deep CNN architecture which is named deep residual learning (DRL-STORM). A residual layer is added to connect two parts and to reduce noise. The first part of the architecture aims to identify redundant information including noise which is then separated from the original raw image by the residual layer. This facilitates molecules localization in the second part of the network. The comparison of two-part architecture (DRL-STORM) and single-part architecture (Deep-STORM) is illustrated in Fig. 3. Different types of blocks in the architectures are highlighted with different colors for distinction. The width of each block is the number of filters in the corresponding convolutional layer. The benefits of using two-part are introducing more parameters, and exploring larger computing power of deep learning from both depth and width, while single-part network cannot.8 The loss function and training data are similar to those of Deep-STORM, and DRL-STORM spends typically 1h in the whole training process.
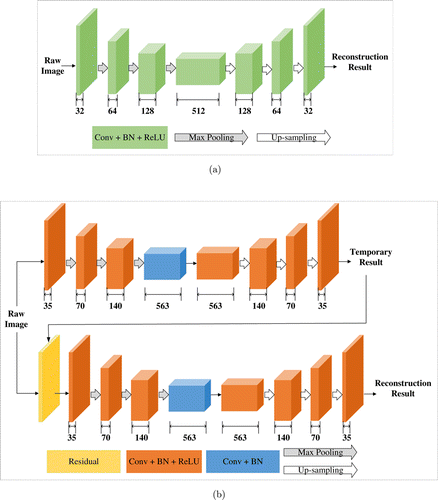
Fig. 3. Comparison of (a) single-part network architecture of Deep-STORM42 and (b) two-part network architecture of DRL-STORM.8 The number below each block refers to the number of features in the representation. Different colors are used to distinct various types of block. (a) The single-part network of Deep-STORM encodes the input raw STORM images and reconstructs super-resolution image in the decoding stage. (b) Two parts of DRL-STORM are connected by a residual layer, which reduces the noise (top part) and localizes high-density molecules (bottom part).
3.2.2. Fluorescence photo-activated localization microscopy
Ouyang et al. present an artificial neural network accelerated PALM (ANNA-PALM), a computational strategy based on deep learning, that reconstructs high-quality, super-resolution images from sparse and far fewer frames of images (two orders of magnitude less) than usually needed.22 Using a small number of frames, this method can improve the temporal resolution without increasing phototoxicity and photobleaching, and preserves high spatial resolution by taking advantage of sparse PALM images as input. The reduction in acquisition time and the improvement in imaging efficiency demonstrate that deep learning can help to achieve faster and high-throughput super-resolution imaging.
The network architecture of ANNA-PALM is a special conditional GAN, consisting of three distinct neural networks. The generator builds on the U-net architecture and produces the reconstructed super-resolution image. The discriminator is a five-layer CNN and provides the adversarial loss. Besides, ANNA-PALM introduces an additional low-resolution estimator that is a four-layer CNN and calculates the low-resolution error map. Dropout layers are employed in the central layers of the generator to prevent the network overfitting.46
There are three distinct loss functions, and each of them is associated to a network. The loss function of the generator penalizes the difference between the generator output and the target image. The difference is defined as a weighted average of the multiscale structural similarity index (MS-SSIM)47 and the norm smoothed by a Gaussian kernel. The loss function of the low-resolution estimator measures the consistency between the low-resolution images produced from the generator and the observed widefield images. This is used to estimate the degree of reliability and highlight potential reconstruction artifacts.22 The loss function of the discriminator is a conditional GAN discriminator loss.
The training data of ANNA-PALM are obtained from standard localization microscopy data. It is worth noting that ANNA-PALM does not necessitate large amounts of training data, but only requires 10 fields of view (FoVs) (of 55m55m each) or even a single FoV of experimental PALM images for training.22 This is accomplished with extensive data augmentation. Unlike Deep-STORM that uses random distributions of molecules as the training data, ANNA-PALM requires the training data with structures similar to those in the images to be reconstructed. Otherwise, the reconstructed results contain errors. Even when applied to data similar to the training images, ANNA-PALM sometimes may produce artifacts. The issue of deceptive artifacts will be further discussed and analyzed in Sec. 4. Training ANNA-PALM from scratch takes on the order of hours to days with a batch size of 1, but retraining can be done in 1h or less when starting from previously trained model.
3.3. Structured illumination microscopy
The fundamental principle of SIM is the computational synthesis of images acquired by shifted illumination patterns. Reconstruction algorithms based on deep learning can be applied to boost SIM’s performance under low-intensity, distorted illumination patterns or challenging SNR conditions.
Qiao et al. present a deep Fourier channel attention network (DFCAN) and its derivative trained with GAN (DFGAN). They can achieve robust reconstruction of SIM images under low SNR conditions and find widespread applications in live-cell imaging over a longer time.48 This suggests that well-designed deep learning models are helpful for SIM in obtaining high-quality super-resolution images, while traditional computational reconstruction is prone to introduce artifacts to the super-resolved images at low fluorescence level. Moreover, this work also provides a comprehensive benchmark for evaluating the fidelity and quantifiability of deep-learning-based super-resolution models, which is important for applying deep learning to enhance the resolution of microscopy images.
The architecture of DFGAN is constructed based on the conditional GAN framework where the DFCAN acts as the generator, and its discriminator is a typical CNN architecture of 12 convolutional layers. DFCAN makes use of the frequency content difference across distinct features in the Fourier domain, rather than structural differences in the spatial domain, enabling the network model to learn the hierarchical representations of high-frequency more precisely and efficiently.48 Fourier channel attention block (FCAB) in each residual group of DFCAN is devised to extract high frequency features.
The loss function of DFCAN is defined as a combination of MSE loss and SSIM loss balanced by a scalar weight , given by
The loss function of the generator of DFGAN is therefore the sum of and discriminative error , i.e.,
The training data are super-resolution and low-resolution image pairs. The former is the reconstructed superior quality SIM image, while the latter is a corresponding wide-field image obtained by averaging raw SIM images. As a result, the training image pairs are well matched and there is no need for registration.48 The authors capture raw SIM images with multimodality SIM systems, and provide an open-access experimental dataset (BioSR) covering wide ranges of SNR levels and biological structural complexities, different levels of excitation light intensity and upscaling factors. Transfer learning has also been employed to facilitate the application of DFCAN and DFGAN to more types of biological structures with a fast training process. The final model of DFGAN takes about 80h to train.
In another application of deep learning to reconstruct SIM data, Jin et al. develop an efficient deep learning-assisted SIM (DL-SIM) reconstruction algorithm that needs fewer frames of raw SIM images and works under low light conditions, thus increasing acquisition speed and reducing photobleaching.15 The authors first employ U-net architecture to achieve comparable resolution with only 3 images (9 or 15 images for conventional SIM), and then connect two U-nets through skip-layer connection to improve reconstruction performance under low laser power and short exposure time. DL-SIM is trained by taking real SIM raw images as the input and corresponding conventional SIM reconstruction results as the ground truth. It takes 2–3 days (2000 epochs) to train the network with 1000 training samples to get a working model for each structure. After transfer learning, the model trained with 200 epochs can produce comparable results. The loss function calculates the pixelwise accuracy without other regularization terms, as follows:
3.4. Three-dimensional (3D) super-resolution microscopy
Since most biological samples are three-dimensional, it is necessary to develop 3D image processing algorithms for analyzing fluorescence microscopic data. There has been research applying deep learning to achieve 3D super-resolution imaging of fluorescent samples,49,50 and the existing methods can also be extended to 3D localization microscopy or 3D SIM reconstruction.48,51
Boyd et al. present a fast molecule localization algorithm (DeepLoco) for both 2D and 3D SMLM data.49 Instead of predicting high-resolution images in Deep-STORM, DeepLoco maps a frame of raw image directly to a collection of molecular locations. The network is conducted based on a standard CNN architecture and is trained with simulated training data. During data generation, the authors use laterally translated versions of an empirically measured PSF and multiple -stacks of different fluorophores, enabling the trained network to be robust to aberrations such as dipole effects.52 Training the neural network takes several hours. A kernel-based loss function is introduced to estimate the squared error of distance between the resulting rendered image and ground truth image in continuous domain.
Wu et al. present a deep-learning-based approach (Deep-Z) that refocuses a single 2D fluorescence image onto 3D surfaces within the sample volume, digitally increasing the depth-of-field of the microscopy without any axial scanning.50 The architecture of Deep-Z is formed by a least square GAN, and the network is trained using accurately matched pairs of fluorescence images acquired at different depths and their corresponding ground truth labels captured at the target focal plane. On average, the training takes 70 h for 50 epochs. The authors use digital propagation matrix (DPM) to computationally correct for aberrations such as sample drift, tilt and spherical aberrations. The least square loss function, rather than cross entropy loss, is adopted for the discriminator to overcome the problem of vanishing gradients,53 and the loss function of the generator combines the traditional content loss (mean absolute error, MAE) and the adversarial loss.
Nehme et al. present a deep neural network (DeepSTORM3D) that is able to localize multiple molecules in three dimensions in densely labeled samples.51 It is not a simple extension of Deep-STORM (2D). Instead, the approach jointly learns the optimal PSF (encoding) and associated localization algorithm (decoding). Two different CNN architectures are used for localizing emitters and for learning a phase mask, respectively, and the networks are trained solely on simulated data. The recovery network training takes about 35 h with a batch size of 4. The learned PSF has a smaller lateral footprint than Tetrapod PSF, which is preferable for minimizing overlap at high densities. The loss function of DeepSTORM3D combines a term of heatmap matching which measures the proximity of the network prediction to the simulated ground truth and a term of overlap measure that provides a soft approximation of the true positive rate in the output image.51
As listed in Table 1, our review covers a large proportion of representative deep-learning-based methods currently available in super-resolution fluorescence microscopy.
| Methods | Architecture | Training data | Loss function |
|---|---|---|---|
| Cross-modality13 | GAN | Experimental data | MSESSIM |
| BCE | |||
| Deep-STORM42 | CNN | Simulated data/Experimental data | Fidelity term regularization |
| DRL-STORM8 | CNN | Simulated data | Fidelity term regularization |
| ANNA-PALM22 | GAN | Simulated data/Experimental data | MS-SSIM loss |
| Consistency loss | |||
| BCE | |||
| DFGAN48 | GAN | Experimental data | MSESSIM |
| BCE | |||
| DL-SIM15 | CNN | Experimental data | Pixel-wise loss |
| DeepLoco49 | CNN | Simulated data | MSE |
| Deep-Z50 | GAN | Experimental data | MAEadversarial loss |
| Least square loss | |||
| DeepSTORM3D51 | CNN | Simulated data | Heatmap matching termoverlap measurement term |
Table 2 firstly summarizes the inference time for one frame of image with different spatial sizes using each method. Deep-STORM and DeepLoco are also compared to conventional methods without using deep learning, implying a speed-up of roughly 1–3 orders of magnitude. In order to show the performance enhancement more informatively and more clearly, we also exhibit the measurements of different metrics in Table 2. Here we only select representative results to quantitatively evaluate the overperformance of each method. For example, peak signal-to-noise ratio (PSNR), normalized root-mean-square error (NRMSE) and SSIM values provided by DL-SIM are calculated for microtubules, and SSIM and RMSE values reported by Deep-Z are from 3D imaging of C. elegans neuron nuclei at location of 5m. More results and analysis can be obtained in the corresponding references. The comparison with the quality assessment of conventional techniques further confirms the gain of using deep learning.
| Methods | Inference time (image size) | Metrics | Performance of Conventional methods |
|---|---|---|---|
| Cross-modality13 | 0.4s | Confocal image | |
| 10241024 pixels | |||
| Deep-STORM42 | 0.011s | % | FALCON54: |
| 512512 pixels | NMSE=61% | ||
| 0.054s | Runtime: 0.338s | ||
| 10241024 pixels | 512512 pixels | ||
| 0.868s | |||
| 10241024 pixels | |||
| DRL-STORM8 | 0.005s | Temporal resolution: 0.5s | |
| 3030 pixels | |||
| ANNA-PALM22 | 1s or less | MS- | PALM image: MS- |
| 25602560 pixels | |||
| DFGAN48 | less than 1s | ||
| 10241024 pixels | MS- | Richardson–Lucy algorithm: | |
| nm | |||
| MS- | |||
| nm | |||
| SIM image: | |||
| MS- | |||
| nm | |||
| Hessian algorithm55: | |||
| MS- | |||
| nm | |||
| DL-SIM15 | Unknown | Low light SIM image: | |
| DeepLoco49 | 0.00005s | Spliner56: | |
| 6464 pixels | RMSE (nm) = 16.126.56 | ||
| z RMSE | RMSE | ||
| RMSE | |||
| Runtime: 0.10101s | |||
| Deep-Z50 | 0.2s | Single fluorescence image: | |
| 512512 pixels | |||
| 1s | |||
| 15361536 pixels | |||
| DeepSTORM3D51 | 0.6s | Matching Pursuit approach57: | |
| 1250750 pixels | Lateral nm | ||
| Axial nm | Lateral nm | ||
| Axial nm |
4. Challenges and Potential Solutions
Despite its prominent successes, the applications of deep learning also encounter challenges and limits. This section discusses the key questions and potential solutions.
4.1. Effects of training data
The amount of training data and the quality of the data are two main aspects affecting the network performance. Insufficient training data can easily lead to overfitting which means that the network fails to generalize to unseen data. Inferior quality training images affect the performance of the network inference. This can be observed from Fig. 4. Even though the input image [Fig. 4(a)] and STED image [Fig. 4(d)] are captured with the same imaging modality, the output image [Fig. 4(c)] of the network trained by using deconvolution of the STED images are worse than the result [Fig. 4(b)] of the network trained with high-quality SIM images. This is related to the fact that the quality of the information in the training examples has an effect on the pixel-to-pixel transformation and the resolution enhancement learned by the network.
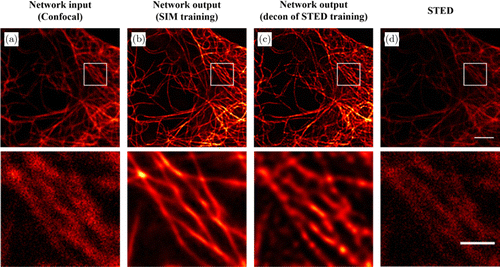
Fig. 4. Comparison of the network output images by the model trained with different training data. (a) A diffraction-limited confocal microscopy image of the microtubules used as input to the network. (b) Super-resolved network inference image by a model trained with high-quality SIM images. (c) Super-resolved network inference image by a model trained with the deconvolution of the STED images. (d) A STED image of the same field of view. The white box regions are shown below at a magnified scale. Scale bars in the first row and second row are 3m and 1m, respectively.
For the problem of lack of data, data augmentation techniques and transfer learning are commonly employed to expand the training dataset. It is worth noting that some operations of data augmentation may not add new information or patterns to the training dataset. In addition, data augmentation should be performed cautiously so as to avoid changing the color or the texture that is associated with the biomolecular information.27 Although different transfer learning strategies can be selected according to the distance of the tasks and domains, it is better to pretrain the network on large collections of high-quality fluorescence microscopy images than on ImageNet. As mentioned in the above section, sufficient training datasets can be produced by simulations. Nevertheless, if we expect the models trained with simulated training data to achieve better performance, then the degradation process in simulation should be as complicated and accurate as that in real applications.58
To obtain high-quality real images for training, it is necessary to design and perform the experiments carefully. Furthermore, the images acquired from different imaging modalities or under different conditions should be taken into account to improve the generalization of the approach. This poses a challenge to the devices and the skills of the technicians. The creation and facilitation of access to large databases of fluorescence microscopic data is always welcomed.
In recent years, meta-learning is proposed to train the network with small amount of simulated training data, enabling the optimal model to generalize to new tasks not seen in the training set.59 Compared to conventional deep learning that heavily relies on abundant training examples, meta-learning is closer to human visual systems that recognize new objects after learning from a limited amount of labeled instances. Another remarkable advantage is that meta-learning can extract and propagate transferable knowledge from a collection of tasks to prevent overfitting and improve generalization.60 Meta-learning-based image reconstruction or resolution enhancement of fluorescence microscopic images is still under-explored and deserves further investigation, mitigating the effect of the training data.
4.2. Issue of reliability
The reliability of the results when applying deep learning in fluorescence microscopy has been questioned. Fake structures and deceptive artifacts may be generated by deep-learning-based methods, which appear plausible and can mislead the subsequent research of biological processes. Some network architectures, such as GANs, are particularly susceptible to hallucinate details.12 Consequently, it is necessary to alleviate this problem and produce reliable images in practical applications of deep learning to fluorescence microscopy. Take ANNA-PALM22 as an example, it improves the reliability of reconstruction results by introducing error maps through the measurement of consistency. The effect of the mismatch between the training data and input data on the network reconstructions is shown in Fig. 5. The network is trained only once and then is applied to the input images [Figs. 5(a), 5(d), 5(g) and 5(j)] that have different mismatch levels with the training data. The corresponding network outputs [Figs. 5(b), 5(e), 5(h) and 5(k)] and error maps [Figs. 5(c), 5(f), 5(i) and 5(l)] of the same area enclosed by black boxes increase for higher mismatch levels, revealing larger inconsistency between the reconstructed super-resolution image and the wide-field image. The error maps or disagreement is helpful for the researchers to determine whether the results are reliable and when the model needs to be retrained with new training data.
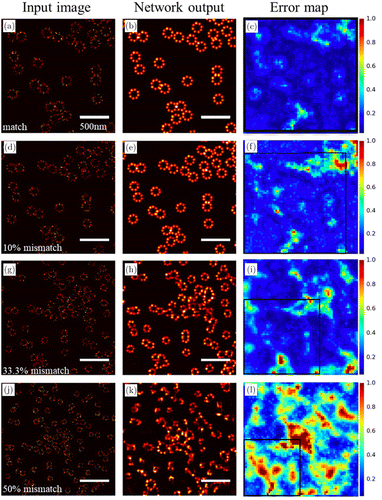
Fig. 5. ANNA-PALM reconstructions and error maps for different mismatch levels between training and input data.22 The first column (a, d, g, j) shows the input images that have different mismatch levels with the training data. The second column (b, e, h, k) shows the corresponding ANNA-PALM network outputs. The third column (c, f, i, l) shows the error maps. The black box encloses the same area as in (a–c). It is clear that the reconstructed images are inferior and the error map values increase for larger mismatch between training data and input data.
Interpreting the “black-box” models is intricate because deep neural networks usually contain complex multilayers and nonlinear interaction components. Fortunately, some studies have investigated or developed the techniques to facilitate the interpretation of the results. For example, Maaten et al. propose a variation of stochastic neighbor embedding (t-SNE), which can visualize the relationships between data by mapping deep network representations to low-dimensional spaces.61 Simonyan et al. explore which parts of the input image are essential for prediction of the output.62 Zeiler et al. analyze the function of intermediate layers to visualize and understand the convolutional networks.63 Lakshminarayanan et al. present a simple and high quality predictive uncertainty estimation using deep ensembles.64 These ideas are suggested to be adapted to fluorescence imaging to quantify and improve the reliability of the results, as has been demonstrated in content-aware image restoration (CARE) networks.24 CARE provides per-pixel confidence intervals and ensemble disagreement scores that can identify image regions where the errors occur.
5. Conclusions
As an advanced data analysis method, deep learning gives a much needed boost to the super-resolution fluorescence microscopy. The representative applications in this field are reviewed in this work. By constructing different network architectures, using simulated or experimental training data and devising various loss functions, current applications take advantage of the computational advances of deep learning to improve the resolution of fluorescence images or to reconstruct super-resolution images with fewer raw data. Although deep learning is still in its infancy for applications in fluorescence microscopy and requires overcoming the limits and hurdles, robust and reliable deep learning methods hold many promises for this field, especially with increasing demand for highly accurate and fast live-cell imaging.
Acknowledgments
This work has been partially supported by the National Key R&D Program of China (2021YFF0502900) and the National Natural Science Foundation of China (61835009/62127819).
Conflict of Interest
The authors declare that there are no conflicts of interest relevant to this article.
References
- 1. , “Beiträge zur theorie des mikroskops und der mikroskopischen wahrnehmung,” Arch. Mikrosk. Anat. 9, 413–418 (1873). Crossref, Google Scholar
- 2. , “Quantitative evaluation of software package for single-molecule localization microscopy,” Nat. Methods 12(8), 717–724 (2015). Crossref, Web of Science, Google Scholar
- 3. , “Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy,” Opt. Lett. 19, 780–782 (1994). Crossref, Web of Science, Google Scholar
- 4. , “STED super-resolved microscopy,” Nat. Methods 15(3), 173–182 (2018). Crossref, Web of Science, Google Scholar
- 5. , “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),” Nat. Methods 3(10), 793–796 (2006). Crossref, Web of Science, Google Scholar
- 6. , “Imaging intracellular fluorescent proteins at nanometer resolution,” Science 313(5793), 1642–1645 (2006). Crossref, Web of Science, Google Scholar
- 7. , “Three-dimensional localization of single molecules for super resolution imaging and single-particle tracking,” Chem. Rev. 117(11), 7244–7275 (2017). Crossref, Web of Science, Google Scholar
- 8. , “Image reconstruction with a deep convolutional neural network in high-density super-resolution microscopy,” Opt. Express 28(10), 15432–15446 (2020). Crossref, Web of Science, Google Scholar
- 9. , “Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy,” J. Microsc. 198, 82–87 (2000). Crossref, Web of Science, Google Scholar
- 10. , “Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics,” Science 349(6251), aab3500 (2015). Crossref, Web of Science, Google Scholar
- 11. , “Deep learning enables structured illumination microscopy with low light levels and enhanced speed,” Nat. Commun. 11, 1934 (2020). Crossref, Web of Science, Google Scholar
- 12. , “Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction,” Nat. Methods 16, 1215–1225 (2019). Crossref, Web of Science, Google Scholar
- 13. , “Deep learning enables cross-modality super-resolution in fluorescence microscopy,” Nat. Methods 16, 103–110 (2019). Crossref, Web of Science, Google Scholar
- 14. , “Efficient image reconstruction of high-density molecules with augmented Lagrangian method in super-resolution microscopy,” Opt. Express 26(19), 24329–24342 (2018). Crossref, Web of Science, Google Scholar
- 15. , “Deep learning enables structured illumination microscopy with low light levels and enhanced speed,” Nat. Commun. 11, 1934 (2020). Crossref, Web of Science, Google Scholar
- 16. , “ImageNet classification with deep convolutional neural networks,” Proc. 25th Int. Conf. Neural Information Processing Systems (NeurIPS), F. PereiraC. J. C. BurgesL. BottouK. Q. Weinberger, Eds., pp. 1097–1105, Curran Associates Inc. (2012). Google Scholar
- 17. , “Fully convolutional networks for semantic segmentation,” in Proc. IEEE Conf. Computer Vision and Pattern Recognition (CVPR), pp. 3431–3440, IEEE (2015), pp. 3431–3440. Crossref, Google Scholar
- 18. , “U-net: Convolutional networks for biomedical image segmentation,” Int. Conf. Medical Image Computing and Computer-Assisted Intervention (MICCAI), N. NavabJ. HorneggerW. M. WellsA. F. Frangi, Eds., pp. 234–241, Springer, New York (2015). Crossref, Google Scholar
- 19. , “Deep convolutional neural network for inverse problems in imaging,” IEEE Trans. Image Process. 26(9), 4509–4522 (2017). Crossref, Web of Science, Google Scholar
- 20. , “Deep learning microscopy,” Optica 4(11), 1437–1443 (2017). Crossref, Web of Science, Google Scholar
- 21. K. Simonyan, A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” preprint, https://arxiv.org/abs/1409.1556 (2015). Google Scholar
- 22. , “Deep learning massively accelerates super-resolution localization microscopy,” Nat. Biotechnol. 36, 460–468 (2018). Crossref, Web of Science, Google Scholar
- 23. , “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell 172, 1122–1131 (2018). Crossref, Web of Science, Google Scholar
- 24. , “Content-aware image restoration: Pushing the limits of fluorescence microscopy,” Nat. Methods 15(12), 1090–1097 (2018). Crossref, Web of Science, Google Scholar
- 25. , Deep Learning: A Practitioner’s Approach, O’Reilly Media, United States of America (2017). Google Scholar
- 26. , “Deep learning,” Nature 521(7553), 436–444 (2015). Crossref, Web of Science, Google Scholar
- 27. , “Deep learning a boon for biophotonics,” J. Biophotonics 13(6), e201960186 (2020). Crossref, Web of Science, Google Scholar
- 28. , “Deep sparse rectifier neural networks,” 14th Int. Conf. Artificial Intelligence and Statistics (AISTATS), G. J. GordonD. B. DunsonM. Dudík, Eds., pp. 315–323, Society for Artificial Intelligence and Statistics (2011). Google Scholar
- 29. , “Interpretation of intelligence in CNN-pooling processes: a methodological survey,” Neural Comput. Appl. 32, 879–898 (2020). Crossref, Web of Science, Google Scholar
- 30. , Deep Learning, The MIT Press, United States of America (2016). Google Scholar
- 31. , “Large-scale machine learning with stochastic gradient descent,” 19th Int. Conf. Computational Statistics (COMPSTAT), Y. LechevallierG. Saporta, Eds., pp. 177–186, Springer, New York (2010). Crossref, Google Scholar
- 32. D. P. Kingma, J. Ba, “Adam: A method for stochastic optimization,” preprint, https://arxiv.org/abs/1412.6980 (2014). Google Scholar
- 33. I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, Y. Bengio, “Generative adversarial networks,” preprint, https://arxiv.org/abs/1406.2661 (2014). Google Scholar
- 34. , “Unrolled generative adversarial networks,” 5th Int. Conf. Learning Representations (ICLR), OpenReview.net, Toulon, France, 2017, pp. 1–25. Google Scholar
- 35. M. Arjovsky, S. Chintala, L. Bottou, “Wasserstein GAN,” Preprint at https://arxiv.org/abs/1701.07875v1 (2017). Google Scholar
- 36. , “VEEGAN: reducing mode collapse in GANs using implicit variational learning,” 31st Conf. Neural Information Processing Systems (NeurIPS), U. V. LuxburgI. GuyonS. BengioH. WallachR. Fergus, Eds., pp. 3310–3320, Curran Associates Inc. (2017). Google Scholar
- 37. , “Mode seeking generative adversarial networks for diverse image synthesis,” 2019 IEEE/CVF Conf. Computer Vision and Pattern Recognition (CVPR), pp. 1429–1437, IEEE (2019). Crossref, Google Scholar
- 38. D. Berthelot, T. Schumm, L. Metz, “BEGAN: Boundary equilibrium generative adversarial networks,” preprint, https://arxiv.org/abs/1703.10717 (2017). Google Scholar
- 39. , “Image-to-image translation with conditional adversarial networks,” 2017 IEEE Conf. Computer Vision and Pattern Recognition (CVPR), pp. 5967–5976, IEEE (2017). Crossref, Google Scholar
- 40. O. Ronneberger, P. Fischer, T. Brox, “U-net: convolutional networks for biomedical image segmentation,” preprint, https://arxiv.org/abs/1505.04597 (2015). Google Scholar
- 41. , “A survey on transfer learning,” IEEE T. Knowl. Data. En. 22(10), 1345–1359 (2010). Crossref, Web of Science, Google Scholar
- 42. , “Deep-STORM: Super-resolution single-molecule microscopy by deep learning,” Optica 5(4), 458–464 (2018). Crossref, Web of Science, Google Scholar
- 43. , “Batch normalization: accelerating deep network training by reducing internal covariate shift,” Proc. 32nd Int. Conf. Machine Learning (ICML), B. FrancisB. David, Eds., pp. 448–456, IMLS (2015). Google Scholar
- 44. , “Imagej2: Imagej for the next generation of scientific image data,” BMC Bioinf. 18, 529 (2017). Crossref, Web of Science, Google Scholar
- 45. , “ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging,” Bioinformatics 30(16), 2389–2390 (2014). Crossref, Web of Science, Google Scholar
- 46. , “Dropout: A simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res. 15, 1929–1958 (2014). Web of Science, Google Scholar
- 47. , “Multiscale structural similarity for image quality assessment,” in 37th Asilomar Conf. Signals, Systems & Computers, pp. 1398–1402, IEEE (2003). Crossref, Google Scholar
- 48. , “Evaluation and development of deep neural networks for image super-resolution in optical microscopy,” Nat. Methods 18, 194–202 (2021). Crossref, Web of Science, Google Scholar
- 49. N. Boyd, E. Jonas, H. P. Babcock, B. Recht, “DeepLoco: Fast 3D localization microscopy using neural networks,” preprint, https://www.biorxiv.org/content/10.1101/267096v1 (2018). Google Scholar
- 50. , “Three-dimensional virtual refocusing of fluorescence microscopy images using deep learning,” Nat. Methods 16(12), 1323–1331 (2019). Crossref, Web of Science, Google Scholar
- 51. , “DeepSTORM3D: Dense 3D localization microscopy and PSF design by deep learning,” Nat. Methods 17, 734–740 (2020). Crossref, Web of Science, Google Scholar
- 52. , “Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking,” Chem. Rev. 117, 7244–7275 (2017). Crossref, Web of Science, Google Scholar
- 53. , “Least squares generative adversarial networks,” 2017 IEEE Int. Conf. Computer Vision (ICCV), pp. 2813–2821, IEEE (2017). Crossref, Google Scholar
- 54. , “FALCON: Fast and unbiased reconstruction of high-density super-resolution microscopy data,” Sci. Rep. 4, 4577 (2014). Crossref, Web of Science, Google Scholar
- 55. , “Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy,” Nat. Biotechnol. 36(5), 451–459 (2018). Crossref, Web of Science, Google Scholar
- 56. , “Analyzing single molecule localization microscopy data using cubic splines,” Sci. Rep. 7(1), 552 (2017). Crossref, Web of Science, Google Scholar
- 57. , “Multicolour localization microscopy by point-spread-function engineering,” Nat. Photonics 10, 590 (2016). Crossref, Web of Science, Google Scholar
- 58. , “Toward real-world single image super-resolution: A new benchmark and a new model,” 2019 IEEE Int. Conf. Computer Vision (ICCV), pp. 3086–3095, IEEE (2019). Crossref, Google Scholar
- 59. , “Model-agnostic meta-learning for fast adaptation of deep networks,” 34th Int. Conf. Machine Learning (ICML), D. PrecupY. W. Teh, Eds., pp. 1126–1135, IMLS (2017). Google Scholar
- 60. W.-Y. Chen, Y.-C. Liu, Z. Kira, Y.-C. F. Wang, J.-B. Huang, “A closer look at few-shot classification,” preprint, https://arxiv.org/abs/1904.04232v2 (2020). Google Scholar
- 61. , “Visualizing data using t-SNE,” J. Mach. Learn. Res. 9, 2579–2605 (2008). Web of Science, Google Scholar
- 62. K. Simonyan, A. Vedaldi, A. Zisserman, “Deep inside convolutional networks: visualising image classification models and saliency maps,” preprint, https://arxiv.org/abs/1312.6034 (2013). Google Scholar
- 63. , “Visualizing and understanding convolutional networks,” 13rd European Conf. Computer Vision (ECCV), D. FleetT. PajdlaB. SchieleT. Tuytelaars, Eds., pp. 818–833, Springer (2014). Crossref, Google Scholar
- 64. , “Simple and scalable predictive uncertainty estimation using deep ensembles,” 31st Int. Conf. Neural Information Processing Systems, U. von LuxburgI. GuyonS. BengioH. WallachR. Fergus, Eds., pp. 6402–6413, NIPS (2017). Google Scholar


