Combination of multi-focus Raman spectroscopy and compressive sensing for parallel monitoring of single-cell dynamics
Abstract
To overcome the low efficiency of conventional confocal Raman spectroscopy, many efforts have been devoted to parallelizing the Raman excitation and acquisition, in which the scattering from multiple foci is projected onto different locations on a spectrometer’s CCD, along either its vertical, horizontal dimension, or even both. While the latter projection scheme relieves the limitation on the row numbers of the CCD, the spectra of multiple foci are recorded in one spectral channel, resulting in spectral overlapping. Here, we developed a method under a compressive sensing framework to demultiplex the superimposed spectra of multiple cells during their dynamic processes. Unlike the previous methods which ignore the information connection between the spectra of the cells recorded at different time, the proposed method utilizes a prior that a cell’s spectra acquired at different time have the same sparsity structure in their principal components. Rather than independently demultiplexing the mixed spectra at the individual time intervals, the method demultiplexes the whole spectral sequence acquired continuously during the dynamic process. By penalizing the sparsity combined from all time intervals, the collaborative optimization of the inversion problem gave more accurate recovery results. The performances of the method were substantiated by a 1D Raman tweezers array, which monitored the germination of multiple bacterial spores. The method can be extended to the monitoring of many living cells randomly scattering on a coverslip, and has a potential to improve the throughput by a few orders.
1. Introduction
Raman spectroscopy is a powerful method that analyzes molecules by probing their vibrational transitions. Taking advantages of its high sensitivity, label-free and nondestructive characteristics, Raman spectroscopy has been widely used in numerous scientific fields such as medical diagnostics, biological investigation, chemical analysis, and environmental monitoring.1,2,3,4 Raman spectroscopy has also been integrated with optical tweezers; the resulted Raman tweezers allow the quantification of biochemical components inside single trapped cells.5,6 Raman tweezers are also easily combined with microfluidic devices, which is very useful for high-throughput cell identification and sorting.7,8 Especially, when artificial intelligence and deep learning are incorporated, Raman tweezers allow improved identification accuracy of microbes.9,10 Moreover, confocal Raman spectroscopy has also frequently been used for monitoring the intracellular dynamics of single cells and for studying the heterogeneity in cell populations.11,12,13
A limitation of traditional confocal Raman spectroscopy is its low efficiency, since it can only analyze one focal point at each time. Many efforts have been devoted to improving the throughput of confocal Raman spectroscopy, including parallel excitation and compressive sensing.14,15,16,17,18,19,20,21,22,23,24 Our group recently combined parallel excitation and compressive sensing to speed up the data acquisition of confocal Raman spectroscopy. We used an array of focused laser beams to excite up to 324 locations on a sample, and organized their scattering in a 2D array on the entry plane of a spectrometer, which then positioned the spectra of different foci on the CCD along both its vertical and horizontal dimensions.25 Multiple spectra were compressed into one spectral channel, which were then recovered by pseudo inversion using their principal components as a prior. We later used the same excitation and detection geometry for single cell analysis and developed a hierarchical sparsity algorithm under the compressive sensing framework, which provided higher reconstruction accuracy and improved signal-to-noise ratios (SNRs).26 However, when applied to monitoring the dynamics of multiple cells, these “noncollaborative” methods ignore the information connection between the cells’ spectra acquired at different time.
In this work, under the same Raman excitation and detection geometry, we developed a collaborative optimization method for demultiplexing the superimposed spectra of multiple biological cells during their dynamics. The method utilized the fact that the spectra of a cell acquired at different time have the same sparsity structure in their principal components. Basing on this a prior, the combined sparsity of the cells’ spectra at different time was penalized when solving the inversion problem. In other words, the time-lapsed spectra of the individual cells were retrieved in a collaborative manner. The method was used to monitor the dynamic germination of multiple trapped bacterial spores. The results indicated that the developed algorithm can accurately retrieve the Raman spectra of the individual cells at all time, and the reconstruction accuracy was much higher than that of the noncollaborative method. The method can be extended to the monitoring of many living cells randomly scattering on a coverslip, and has the potential to improve the throughput by a few orders.
2. Materials and Methods
2.1. Materials
The biological cells involved in this study were yeast cells (strain of FB86) as well as spores of Nosema bombycis (Nb) and Bacillus thuringiensis (Bt, strain of AS1.1754). The yeast strain of FB86 was isolated from sea mud and cultured at 26∘C in YPD medium (10g/L yeast extract, 20g/L peptone, and 20g/L dextrose) for 48h. The yeast cells were harvested and purified by centrifugation at 5000rpm for 8min. The cells were then washed three times with sterile water. The Nb spores used in this study were propagated and purified from infected silkworm larvae. Leaves that were artificially contaminated using a spore suspension (∼ 1 × 107spores/mL) were fed to third instar host larvae. Silk glands and midguts of fifth instar larvae were collected and homogenized with a glass homogenizer. Spores were then extracted from the crude spore suspension and purified using sucrose density gradient centrifugation. The Bt spores were activated on Luria broth (LB) agar plates containing 10g/L peptone, 5g/L yeast extract, and 10g/L NaCl (pH 7.4) for 24–36h. A selected single colony was inoculated in LB broth and cultured with agitation at 200rpm for 12h. One percent of the bacterial culture was then inoculated in sporulation medium and incubated with agitation at 200rpm for 48–60h. Spores were harvested and purified by centrifugation at 5000rpm for 10min and washed 10 times with sterile water. The purified spores and cells were suspended in sterile water and stored at 4∘C until use. Germination of spores of Bt was triggered by 30mM external CaDPA (calcium-dipicolinic acid) at room temperature.27,28,29
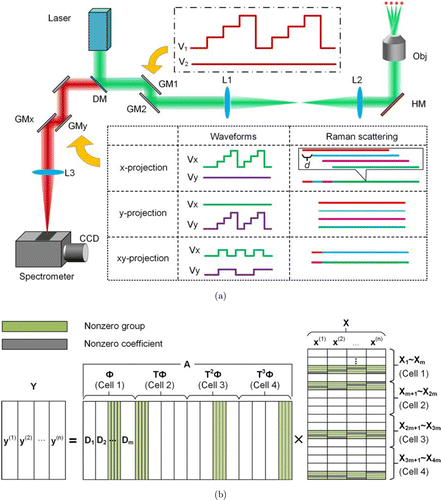
Fig. 1. Compressive multi-focus Raman spectroscopy for parallel monitoring of biological dynamics in multiple cells. (a) Experimental setup. Abbreviations: DM, dichroic mirror; GM, galvo-mirrors; L, lens; HM, hot mirror; Obj, objective. Raman scattering from different cells is represented by different colored stripes. (b) Collaborative hierarchical sparsity model for spectral demultiplexing in the x-projection mode. An example of four cells analyzed by a 4-foci Raman tweezers array is shown.
2.2. Experimental methods
A lab-made confocal Raman microscope was used to monitor the biological dynamics of single cells. The Raman excitation source used in this work was a diode laser at 785nm (Sacher Lasertechnik, TEC-510-0785-1000). The laser beam was introduced into an inverted microscope (TE2000U, Nikon) where it was focused by a high-numeric-aperture (high-NA) objective (100 × oil-immersed, NA of 1.3) to excite the Raman scattering of a single cell. In order to parallelize monitoring of the dynamics in multiple biological cells, an array of focused laser beams are required. For this purpose, a pair of galvo-mirrors (GM1 and GM2, GVS002, Thorlabs, Inc.) were placed at the conjugate plane of the back aperture of the objective, which were driven by a data acquisition card (DAQ, PCIe-6353, National Instruments). By rapidly scanning the galvo-mirrors with staircase waveforms (250Hz), multiple focused laser beams were produced at the focal plane of the objective in a time-sharing manner.14 Figure 1(a) shows the example waveforms (in the dot-dashed rectangle box) that generate a 1D array of focused beams targeting four individual biological cells. In the similar manner, we can generate more complex excitation pattern, and multiple cells scattering on a coverslip that can be excited simultaneously by random accessing scanning.18 The backward-propagating Raman scattering from the cells was collected by the same objective, returning to the galvo-mirrors where it was descanned. The descanned scattering light was seperated from the excitation beam by a dichroic mirror (DM, LPD02-785RU-25, Semrock) and was then focused by a lens (L3 in Fig. 1(a)) onto the entrance of a spectrometer (Acton SP2300i, Princeton Instrument). The spectrometer was equipped with a CCD camera (ProEM, Princeton Instrument) that recorded the dispersed Raman scattering.
To demultiplex the Raman spectra from the multiple cells, another pair of galvo-mirrors, GMx, and GMy, were placed at the back focal plane of the lens L3. When the waveforms applied on them were synchronized with the multi-focus generation, the scattering of different cells were project onto different locations on the entry plane of the spectrometer. When a staircase waveform was applied on GMx, it discretely positioned the scattering of different cells along the horizontal direction (the dispersion direction of the grating). As a result, the spectra of different cells were horizontally displaced with a constant step size of d = 25 pixels on the CCD, and were eventually mixed into a single overlapped spectral stripe (see the x-projection mode shown in Fig. 1(a)), as the exposure time (17s) was much longer than the scanning period (4ms). By binning the spectral stripe over 5 pixels along the vertical direction, a single mixed spectrum was obtained. In this way, the Raman spectra of multiple cells were recorded simultaneously in a single spectral channel, and the single-cell analysis throughput was improved by a factor depending on the number of foci. When the same waveform was applied on GMy, the spectra of different cells were projected onto different row pixels of the CCD, resulting in multiple spectral stripes (see the y-projection mode in Fig. 1(a)). The space between the adjacent stripes was 10 pixels, which was larger than the stripe width (3 pixels in full-width-at-half-maximum). By binning the individual stripes over 5 pixels, spectra of the individual cells were obtained without crosstalk.
In our proposed spectral compressive scheme, the spectra are acquired in xy-projection mode, as shown in Fig. 1(a), which positions the spectra of multiple cells on a CCD along both the horizontal and vertical directions, and thus has much higher throughput of single-cell analysis. In the xy-projection mode, multiple well-seperated spectral stripes were formed on the CCD, and each was a mixture of many different spectra. Since there was no cross talk between the different stripes, the method for demultiplexing the mixed spectra in each stripe was the same as that in the x-projection mode. Thus, for simplicity, our discussion on spectral reconstruction was restricted to the x-projection mode in this work, and the developed method is easily generalized to the xy-projection mode. We also point out that, as the multi-focus generation and the scattering projection are decoupled from each other via descanning and re-scanning of the scattering, the organization pattern of the cells as well as the projection mode can be arbitrary.
In order to monitor the biological dynamics in these cells, their Raman spectra were continuously acquired in both x- and y-projection modes. Without altering the waveforms for multi-focus generation, the two acquisition modes were alternated by switching the waveforms on GMx and GMy every 20s. Each time the waveforms were switched, a TTL pulse was generated by the DAQ card, which triggered the spectrometer’s CCD to start integration. In either detection mode, the average laser power of each focus was 10mW, and the integration time was 17s, which was less than the time interval between the successive modes and thus left enough time for data transferring. The time-lapsed spectra acquired in the x-projection mode were fed into our collaborative hierarchical Lasso algorithm (see below) such that the spectra of the individual cells can be reconstructed at all time intervals. The time-lapsed spectra obtained in the y-projection mode were used as the ground truths to substantiate the accuracy of our demultiplexing method.
2.3. Spectral demultiplexing algorithms
We developed a collaborative hierarchical sparsity model under a compressive sensing framework to demultiplex the mixed spectra acquired in the x-projection mode. This model constructs the spectra of the individual cells from a collection of eigenspectra that are obtained by principal component analysis (PCA) of the training data. Assuming that the cells under investigation are possibly from m cell populations, we randomly pick up a number of cells (e.g., 50 cells) in each cell population and obtain their Raman spectra by single-trap Raman tweezers. PCA analysis is independently applied to the training spectra from each cell population. We then obtain m sub-dictionaries (sets of principal components), namely D1– Dm. When these sub-dictionaries are combined into a larger dictionary Φ, Φ=[D1|D2|…|Dm] (see Fig. 1(b)), the spectrum, s, of any cell in the system can be represented by a combination of the eignspectra in this dictionary, s=Φf, where f is the corresponding coefficient vector. The contributions to a single cell’s spectrum are primarily from only one sub-dictionary, as indicated by the green-shaded regions in Fig. 1(b), since a cell is from only one cell population. Within this sub-dictionary, only a few principal components are dominant, and thus only a few elements in the coefficient vector are nonzero, see the gray-shaded regions in Fig. 1(b).26 For this reason, if the coefficient vector is partitioned into m groups such that each group is corresponding to a sub-dictionary in Φ with the same index, it will exhibit hierarchical sparse structure, i.e., only one group is active for each cell, and only a few elements in this group are nonzero, see the green- and gray-shaded regions on Fig. 1(b). For illustration simplicity, we consider a case that four biological cells from m cell populations are targeted by an array of focused laser beams; their Raman spectra are continuously acquired in the x-projection mode at n different time intervals, resulting in n multiplexed spectra, y(1)−y(n), see Fig. 1(b). Each spectrum is a sum of the spectra of the four cells after translation, according to the x-projection scheme. The multiplexed spectrum acquired at the jth time point, y(j), then can be written as follows :
Rather than solving the linear inversion problem, Eq. (1), independently at each moment,25,26 we choose to jointly solve the inversion problem to the spectral sequence (also shown in Fig. 1(b))
3. Results and Discussion
We demonstrated the feasibility of the developed method and algorithm by monitoring the dynamic germination of multiple bacterial spores with a 1D Raman tweezers array. In this study, Bt spores were mixed with yeast cells, which were then exposed to external CaDPA that triggers spore germination by activating the cortex-lytic enzyme CwlJ, which initiates degradation of the spore’s peptidoglycan cortex and the subsequent germination events.27,28,29 In detail, a sample chamber filled with 30mM CaDPA solution was placed under the microscope, and a drop (5L, cells/ml) of the mixed cell suspension was injected into the chamber. Four cells in the solution were randomly captured by the laser tweezers array, ignoring their sizes and shapes. The tweezers array was generated in a time-sharing manner, as described in Sec. 2, and the average laser power for each trap was 10mW. As shown in Fig. 2(a), one yeast cell and three Bt spores were trapped, and they were organized in a 1D array pattern and separated by 3m from each other. It took less than 1min to load the four cells into the tweezers. The dynamic responses of these cells to the external CaDPA was monitored by Raman scattering, which was continuously acquired in both the -projection mode (our compressive detection scheme) and the -projection mode (to obtain the ground truths for verification purpose), as detailed in Sec. 2. The integration time for each acquisition mode was 17s, and the two acquisition modes were switched every 20s. We should point out that, during the switching of the acquisition mode, the waveforms on GM1 and GM2 were not interrupted, therefore, the four cells stably stayed in their traps. After 24min, we released the cells and acquired the background spectra in both the modes. The background spectra were smoothed using a Savitzky–Golay filter with a window size of 3, which were then subtracted from the corresponding time-lapsed spectra of the cells.
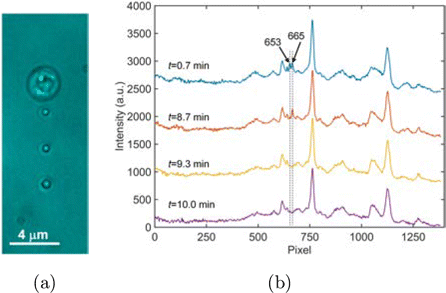
Fig. 2. Monitoring dynamics of four biological cells using a Raman tweezers array and compressive sensing. (a) Bright-field images of four cells optically trapped in 30mM CaDPA solution by a 1D laser tweezers array. From top to bottom: a yeast cell and three Bt spores. (b) Multiplexed Raman spectra of the four cells acquired at different time in the -projection mode. Baselines are shifted for display purpose.
The time-lapsed spectra of the four cells acquired in the -projection mode are shown in Fig. 2(b). Obviously, the peaks centered at the pixels of 653 and 665 disappeared during the dynamic course. However, the assignment of these peaks to the individual cells as well as their wavenumbers were not clear, since the spectra of the different cells were translated and mixed together in the -projection mode. We then applied the C-HiLasso algorithm to retrieve the spectra of the individual cells at different time intervals. As required by the algorithm, we built the dictionary A from polystyrene beads and three cell populations, i.e., yeast cells, Bt, and Nb spores. From each population, we used single-trap Raman tweezers (without scanning the galvo-mirrors in Fig. 1(a) to randomly trap 50 particles and acquired their Raman spectra for PCA analysis). The laser power for training data acquisition was 40mW, and the integration time was 20s for yeast cells, Bt and Nb spores, but was 2s for the polystyrene beads since its Raman scattering was much stronger. The principal components of the four populations were then used to build the dictionary A, as detailed in Sec. 2. The mixed spectra acquired at different time intervals were combined into the matrix Y, and Eq. (3) was solved to obtain the spectra of the four cells at all-time intervals. As shown in Fig. 3, the spectra retrieved using the C-HiLasso algorithm agreed very well with the ground truths that were obtained in the -projection mode, as detailed in Sec. 2. For the yeast cell, the dominant peaks at 1151cm and 1511cm are assigned to the elevated levels of carotenoid in this strain,33 and the small peaks at 1004cm and 1443cm belong to phenylalanine and lipid, respectively.34 For the Bt spores, the peaks at 658, 824, 1017, 1395, and 1572cm are from CaDPA which resides in spores’ core, and the broad band at around 1655cm is associated with the protein amide I.33 We also can see that the retrieved spectra had higher SNR than those acquired in the -projection mode. Two different sources contributed to this improvement. One was that the optimization algorithm rejected the noisy principal components by sparsity penalty. Another was that the SNR of the superimposed spectra was higher, as the signals from different cells were summed up into a single spectrum in the -projection mode, while the dark noises and read-out noises were the same as those in the -projection mode.
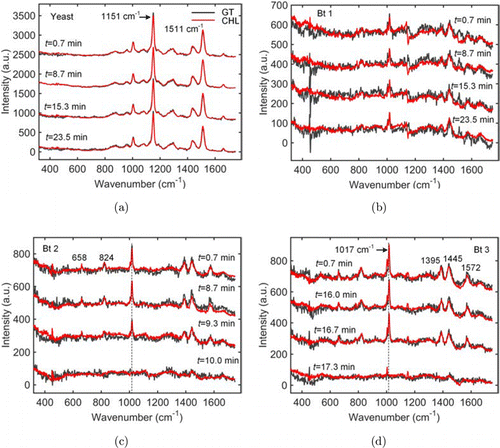
Fig. 3. Retrieved spectra of the individual cells at typical time intervals using the C-HiLasso algorithm. (a) Retrieved spectra (red curves) of the yeast cell at different time as well as its ground truths (black curves). Abbreviations: GT, ground truth; CHL, C-HiLasso. (b)–(d) Retrieved spectra of three Bt spores at different time as well as their ground truths.
It can be concluded from the time-elapsed spectra that, the yeast cell did not respond to CaDPA, and its spectra almost did not change. Moreover, although external CaDPA can trigger germination of bacterial spores, the individual Bt spores in a population behaved very differently when exposed to 30mM CaDPA. While the spores Bt 2 and Bt 3 germinated and released their internal CaDPA at 10.0 and 17.3min, respectively, the spore Bt 1 did not germinate until 23.5min, as indicated by the Raman bands associating to the CaDPA. By using our spectral compressive acquisition scheme, we were able to monitor the dynamics of four cells within 24min using only one spectral acquisition channel, which, however, would take 96min using traditional confocal Raman spectroscopy, since it can analyze only one cell at a time. The improvement in the throughput will be more significant if a laser-beam array with more foci, and if the biological processes in the cells are very long.
As a comparison, we also applied the HiLasso algorithm to demultiplex the time-lapsed mixed spectra, which independently solved the linear inversion problem Eq. (1) at each time point.26 Figure 4 shows the spectra of the individual cells retrieved by the two algorithms at 6min. It is obvious that the spectra reconstructed by the C-HiLasso algorithm provided much higher fidelity than did the HiLasso algorithm. Especially, two dips at 1151cm and 1511cm appear in the Bt spores’ spectra reconstructed by the HiLasso algorithm, which, however, do not show up in those reconstructed by the C-HiLasso algorithm. These dips coincide with the two dominant peaks in the yeast cell’s spectrum, and the collaborative optimization of the inversion problem to the spectral sequence precluded these wrong components. We should point out that, although only the results obtained at one typical time point are shown, the same conclusion was applied to the spectra acquired at other time intervals.
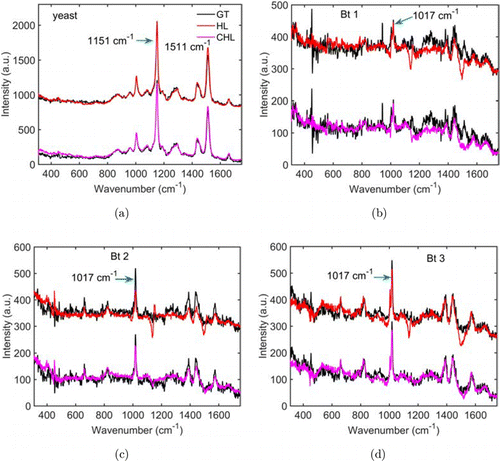
Fig. 4. Spectral demultiplexing of four optically trapped cells acquired at t = 6min using different algorithms. (a)–(d) Spectra of the four cells reconstructed using different algorithms as well as their ground truths. Abbreviations: GT, ground truth; HL, HiLasso and CHL, C-HiLasso.
In addition to providing the retrieved spectra, we also monitored the peak intensities of the characteristic Raman bands of these cells, in order to demonstrate that the C-HiLasso algorithm can quantify single-cell dynamics with higher accuracy. For the yeast cell, we chose to monitor the band at 1151cm (related to carotenoid in the cell) that had maximum SNR. For the Bt spores, we monitored the Raman bands corresponding to the internal CaDPA, which was released from the cores during spore germination. Among all Raman bands related to the CaDPA, the band at 1017cm was mostly dominant, and its peak intensities were used to quantify the germination. Figure 5 shows that the Bt spores 2 and 3 germinated within 24min after the exposure to 30mM external CaDPA, and their internal CaDPA began to release at 9.3 and 16.7min, respectively. Although their lag time to start CaDPA release was different, these spores spent almost the same amount of time (less than 1.5min) to complete their CaDPA release, coinciding to our previous observation.35 It is obvious that, by comparing the reconstructed spectra to the ground truths, the C-HiLasso algorithm quantified the CaDPA contents in the Bt spores with higher accuracy than did HiLasso. It is interesting that we also saw the slow decrease of the carotenoid band in the yeast cell, which may be due to photobleaching. While the intensities of this band obtained by the C-HiLasso algorithm almost followed the same tendency as that obtained in the -projection mode, the results obtained by the HiLasso algorithm were not very stable, and the quantification errors were pretty big at some time points. Therefore, the C-HiLasso algorithm that solved the inversion problem in a collaborative manner outperformed the HiLasso algorithm when applied to quantifying the dynamics of multiple biological cells.
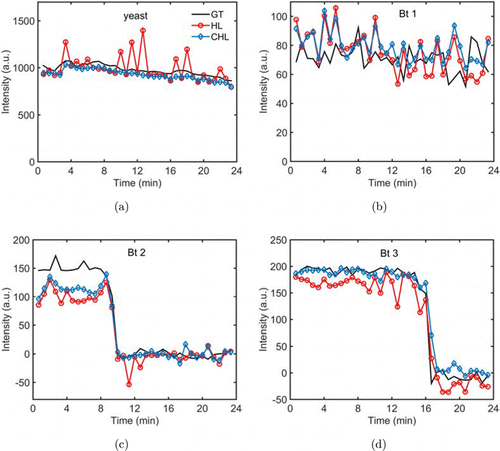
Fig. 5. Peak intensities of the Raman bands indicated by the arrows in Fig. 4 as the function of time. (a) Peak intensities of the band at 1151cm in the spectra of the yeast cell. (b)–(d) Peak intensities of the band at 1017cm in the spectra of the three Bt spores, respectively. Different algorithms were used to retrieve the spectra of the individual cells acquired in the -projection mode, and the peak intensities were calculated by 5-point average. The peak intensities obtained in the -projection mode are also present as the ground truths (black curves). Abbreviations: GT, ground truth; HL, HiLasso, and CHL, C-HiLasso.
4. Conclusions
In summary, we have proposed a technique that allows accurate quantification of the intracellular dynamics of multiple biological cells by confocal Raman spectroscopy. This technique inherited the same excitation and detection geometry reported in our previous work.25 Multiple biological cells are simultaneously excited in a time-sharing manner by rapidly scanning a focused laser beam. After descanning and rescanning, their Raman scattering is organized in a 1D or 2D array on the entry plane of a spectrometer, corresponding to the -, -, or xy-projection modes. In either - or xy-projection mode, the spectra of multiple cells are translated and compressed into a single acquisition channel, which provides increased throughput, shorter readout time, and higher effective SNRs. We have previously demonstrated that, by decomposing their spectra into a serial of principal components, the mixed spectra acquired in these two modes can be demultiplexed by solving the inversion problem under a compressive sensing framework.26 We proposed in this work that the spectra of a cell during its dynamics have the same sparsity structure and demonstrated that, when this a prior was used to solve the inversion problem to the whole spectral sequence, more accurate recovery of the individual spectra was obtained.
As the excitation and detection geometry are decoupled from each other by descanning and re-scanning of the Raman scattering, the organization of the cells does not have to be regular, and the projection mode can be arbitrary too. Therefore, the developed technique can be used to monitor the dynamics in a large number of cells scattering on a coverslip,36 which is more practically applicable than a Raman tweezers array as it does not have to take long time to load many cells into the individual foci. Although the feasibility of this technique was demonstrated by -projection mode, the extension to xy-projection mode is very straight, as the different stripes formed by vertical projection are well-separated as long as their space is larger than their width. For the above reasons, when the xy-projection mode is used for monitoring the cells scattering on a cover slip, the throughput will be improved significantly. The number of cells that can be simultaneously analyzed by this technique is limited by a few factors, including the total laser power, the row and column numbers of CCD pixels as well as the spectral stripes’ width. As the width of the spectral stripe is 5 pixels in our system, a CCD camera with 400 row pixels can record 80 stripes. In each stripe, the translation step size of the individual spectra should be larger than the spectral resolution of the system (about 5 pixels, or equivalently 6cm, and thus the compression of 20 spectra into one stripe only wastes 100 pixels, as comparing to a total of 1600 column pixels of the CCD. Therefore, theoretically, the throughput can be improved by three orders as compared to traditional confocal Raman spectroscopy.25 The technique and algorithm developed in this work are expected to be widely applicable to monitoring of single cell dynamics in a label-free and nondestructive manner.
Acknowledgments
This work was supported by the National Key R&D Program of China (2019YFC1605500, 2018YFF01011700), the National Natural Science Foundation of China (21973111), Guangxi Natural Science Foundation (2017GXNSFAA198029), and Scientific Development Fund of Guangxi Academy of Sciences (2018YFJ403). Zhenzhen Li and Xiujuan Zhang contributed equally to this work.


